Biomedical Engineering Reference
In-Depth Information
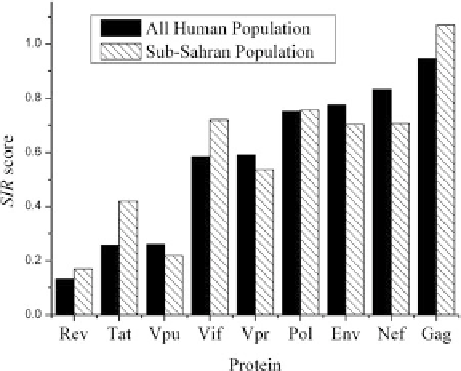
Fig. 6
Average SIR score
of HIV-1 genes. The average
SIR score was computed for
two possible populations: the
all human population and the
sub-Saharan population. The
genes were ordered according
to their score in the all human
population
The SIR results presented up to now represents the average effect on the
population of mutations. These results may be affected by the HLA allele usage
in the population, or by sampling effects on the HLA usage of HIV carriers.
We have thus repeated the analysis using a smaller number of sequences, where
the host HLA serotyping is known. These sequences are the aggregated results of all
HIV-1 clad B cohorts available in the LANL database. We computed for these HIV
sequences the SIR scores in the HLA allele of their host, and used protein/HLA
combinations for which at least ten sequences were available, to obtain reliable
averages. As was observed in the average analysis, Tat and Rev systematically have
low SIR scores, while Gag has high ones (Fig.
6
). Interestingly, when comparing
the SIR of HIV genes on the HLA of their host with the average SIR score of
all sequences (i.e. serotyped and un-serotyped sequences) of the same gene on the
same HLA allele, the weighted average of the difference is significantly negative
for the regulatory genes Tat and Rev and significantly positive for Gag (
R
=
0
.
2,
0001) (Fig.
7
). One can thus clearly see from multiple angles a positive
selection of epitopes in Gag and a negative selection of such epitopes in regulatory
genes in hosts with matching HLA.
The SIR score only provides statistical information. In order to translate these
results to the properties of specific epitopes, we tracked the HLA-A*0201 Gag,
Pol and Tat epitopes over the last 30 years. HLA-A*0201 is the most frequent
allele, and a large number of experimental epitopes were tested on this allele. Tat
has no epitopes in the ancestral sequence and only two epitopes (QPLQIVVIV
and VPIAIVKSV) in all early sequences; both appear in a single sequence, and
disappear over time. Some transient epitopes appear, but only briefly. Not only
do the epitopes themselves disappear, but their position in the genome is highly
mutated. By the end of our sampling it is completely different (only 30 % similarity).
Interestingly, once the epitopes disappeared, this position remained conserved. Rev
evolution is very similar to Tat, showing a systematic elimination of the epitopes.
p
<
0
.
Search WWH ::

Custom Search