Biomedical Engineering Reference
In-Depth Information
1
G1 sim
SG2M sim
G1 data
SG2M data
0.9
0.8
0.7
0.6
0.5
0.4
0.3
0.2
0.1
0
0
5
10
15
20
25
30
time (h)
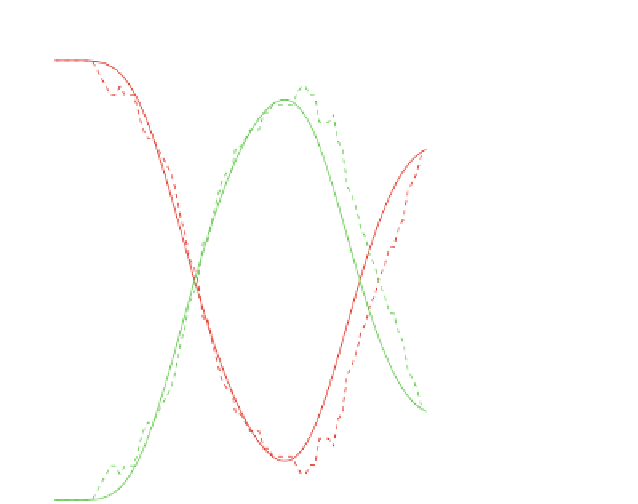
Fig. 2
M
(
green
or
light grey
) from biological data (
dashed line
) and from numerical simulations (
solid line
).
Our model results in a reasonably good fit to biological data
Time evolution of the percentages of cells in the phases
G
1
(
red
or
deep grey
)and
S
/
G
2
/
the random variable corresponding to the time spent in
G
1
and
S
M
.This
enabled us to determine the expression of the transitions rate according to the
formula (
22
). We then compared the solutions of the system with cell recordings
that had previously been synchronised “by hand”, i.e., all recordings were artificially
made to start simultaneously at the beginning of
G
1
phase. The result is shown on
Fig.
2
. Note that using an inverse problem method—see Sect.
5
—instead of ours
could have consisted here in determining the parameters of the model, i.e.,
K
i
→
i
+
1
transition functions, by minimising an
L
2
distance between this experimental data
curve and a theoretical, parameter-dependent curve representing these data.
/
G
2
/
7.5
Optimising Eigenvalues as Objective and Constraint
Functions
We then used combined time-independent data on phase transition functions,
obtained from experimental identification of the parameter functions
K
i
→
i
+
1
(
t
,
x
)=
κ
i
(
x
)
in the uncontrolled model, with cosine-like functions representing the periodic













































Search WWH ::

Custom Search