Biomedical Engineering Reference
In-Depth Information
0.86
0.90
0.84
0.85
0.82
0.80
0.80
0.75
0.78
0.70
0.76
0.74
0.65
0
20 000
40 000
60 000
80 000
100 000
0
20 000
40 000
60 000
80 000
100 000
Sample Size
Sample Size
0.95
0.90
0.85
0.85
0.80
0.80
0.75
0.75
0.70
0.65
0.70
0
20 000
40 000
60 000
80 000
100 000
0
20 000
40 000
60 000
80 000
100 000
Sample Size
Sample Size
Fig. 10.
Sensitivity and PPV of prediction as a function of sample size obtained by considering
100 distinct tRNA sequences from [15]. Plots correspond to those of Fig. 9.
Ta b l e 2 .
Sensitivity and PPV of predicted foldings for 100 distinct tRNA sequences from [15]
(a)
Accuracies derived with
leading prediction tools.
Tool
(b)
Accuracies obtained with heuristic sampling.
Tabulated are the best accuracies (maximizing the
sum of sensitivity and PPV) as observed for one
of the considered sample sizes.
Strategy
Sens. PPV
CONTRAfold
80
71
W
exact
Time
Sens. PPV S. size
PPfold
59
84
∞O
(
n
3
) 88
Conventional
84 1000
RNAshapes
66
60
O
(
n
2
) 88
30
84 60000
Sfold, mfe
64
57
O
(
n
2
) 85
−
1
84 86000
Sfold, ec
61
64
∞O
(
n
3
) 89
Alternative
85 2000
UNAFold
64
59
O
(
n
3
) 87
30
85 19000
ViennaRNA
64
57
O
(
n
2
) 83
−
1
83 73000
several leading RNA secondary structure prediction tools (on the basis of the same
tRNA sequences, as collected in Table 2(a)). Despite the rather simple structure of tRNA
molecules (short sequences with comparably low structural variety), these first results
anyhow indicate the validity of the proposed heuristic sampling approach. Notably, our
novel sampling strategy seems to produce more accurate results than the common one
for particular settings, for instance in case of exact preprocessing, although it was orig-
inally designed to fit especially well with our approximative variants.
In summary, the presented results are quite encouraging, but undoubtably more reli-
able empirical studies in connection with other classes of RNAs (e.g., the ones consid-
ered in [4] for exact preprocessing in combination with the common sampling strategy)
and further prediction selection schemes (e.g., centroids) are required for evaluating our
heuristic method(s) and for identifying potential sites for improvement.
6
Conclusions
The major advantage of the presented approximative method is that it is more efficient
than all other modern prediction algorithms (implemented in popular tools like Mfold [6],
ViennaRNA [8], Pfold [10], Sfold [2] or CONTRAfold [16]), reducing the worst-case
time complexity by a linear factor, such that the time and space requirements are both
bounded by
(
n
2
)
. However, a potential drawback lies in the observation that the overall
O











































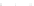


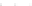









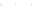
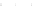



































































Search WWH ::

Custom Search