Biomedical Engineering Reference
In-Depth Information
and b.
45
The analyses of peptides (PICS: proteomic identification of protease
cleavage sites and native substrates (TAILS: terminal amine isotopic labeling of
substrates has allowed for the identification of more than 1000 cleavage sites for
each enzyme.
43-47
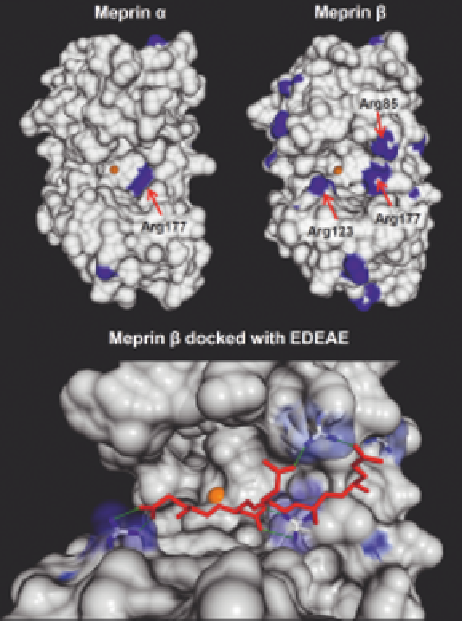
The specificity for negatively charged amino acid residues in
P1
0
was between 50 and 70% for meprin a and b, respectively. This is struc-
turally based on a conserved positively charged residue (Arg177) within the
active site cleft (Figure 2.3A,B). Interestingly, meprin b was even capable of
cleaving within peptides consisting exclusively of negatively charged amino
acids (Figure 2.3C), thereby revealing a unique specificity reflected by sub-
strates such as procollagen III, the amyloid precursor protein (APP), inter-
leukin(IL)-1b, and prokallikrein 7.
34,48-50,54
A
B
C
Figure 2.3
Structural properties of the catalytic domains of human meprins.
Homology surface models of the catalytic domains of human meprin a (A)
and b (B) were computed based on the crystal structure of BMP-1
(3EDG), using Chimera and SwissModeler.
55,56
Positively charged resi-
dues are displayed in blue and the zinc in orange. Amino acids determining
the cleavage specificity are indicated by red arrows. The substrate EDEAE
(red) was docked into the active site cleft of meprin b exhibiting ionic
interactions (green lines) between corresponding residues (C).