Biomedical Engineering Reference
In-Depth Information
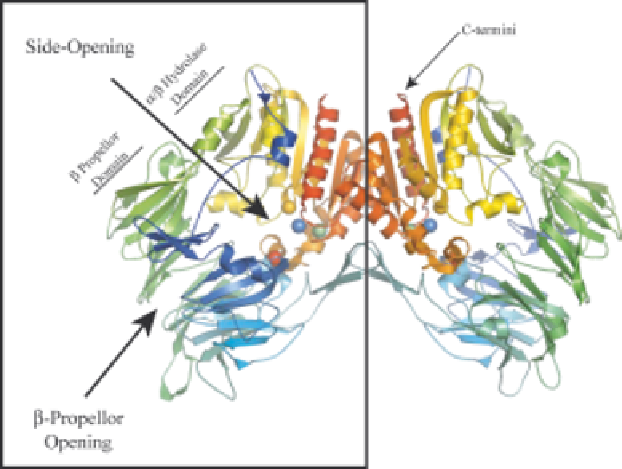
Figure 1.3
Dimeric structure of human DP4 demonstrating entry to the active
site. The catalytic triad residues are displayed as spheres; Red
¼
Ser630;
Green
¼
Asp708 and Blue
¼
His740. The two Glu residues at 205 and
206 that interact with the substrate are displayed as yellow spheres. The
structural representation was generated using PyMOL with structural
coordinates (PDB code 1N1M) from Rasmussen et al.
15
most easily accessible route.
15-17
It is possible that entry and exit of substrates
occur via different routes.
17
Structural resolution of DP4 in complex with
residues 1-10 of its substrate, NPY, provided evidence for preferred entry of
substrates via the large side-opening.
17
Furthermore, docking experiments of
DP4 with aprotinin indicated that the aprotinin N terminus could enter the
active site only via the side-opening.
140
The large cavity is negatively charged
and would most likely attract the positively charged N-terminus of DP4 sub-
strates into the active site.
15
DP4 and FAP structures reveal that the pair of
conserved glutamate residues that are required for enzymatic activity, (Glu
205
,
Glu
206
) in DP4 and (Glu
203
, Glu
204
) in FAP, aid in catalysis due to their
negative charge recognizing the positively charged N-termini of substrate
peptides.
13,139
Resolution of the FAP crystal structure revealed a similar
proposed route of entry to the active site, via one of the two openings within the
b-propeller domain.
13
One of the main requirements for DP4 catalysis to occur
is for a free, unblocked, protonated a-amino group to be present in the P1
position.
141,142
In this regard, FAP differs from DP4 due to its ability to
function also as an endopeptidase. Due to the restricted size of entry into the
active site, it is thought that only elongated peptides or unfolded or partly
unfolded protein fragments can reach the active site.
15
Furthermore, only small