Biomedical Engineering Reference
In-Depth Information
B
A
SMIPP-S-I1
SMIPP-S-D1
Y200
A
Y202
A
D
N
H
Q
Q190
C
SMIPP-S-I1
Q191*
V189
V195
Y200
P113
Q147*
L112
I222
A
A146
D
180˚
H
P13
I226
L34
A110
K108
SS1
D103
L102
123456789
Average
Variable
Conserved
Insufficient data
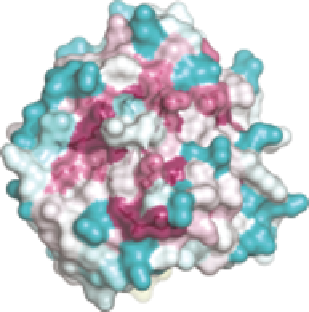
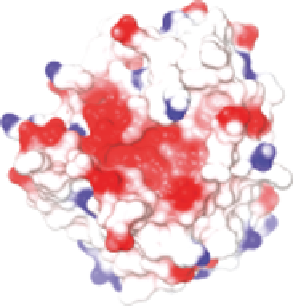
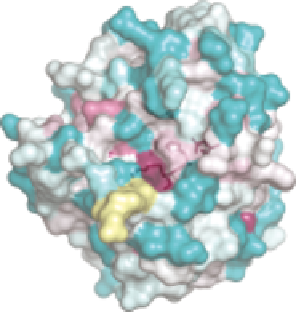
Figure 10.4
Molecular surfaces of (A) SMIPP-S-I1 and (B) SMIPP-S-D1 colour-
coded according to electrostatic potential (red
¼
negative, blue
¼
positive). (C) Sequence conservation of all 33 SMIPP family members
mapped onto the molecular surface of SMIPP-S-I1. Left and right views
are orthogonal. Highly conserved surface-exposed residues, as well
as catalytic triad residues are shown as sticks and labelled. Predicted
N-glycosylation sites are labelled with an asterisk. (D) Sequence con-
servation mapped onto SMIPP-S-D1, as for (C). (E) Structural super-
position of SMIPP-S-I1 and SMIPP-S-D1, both colour-coded according
to sequence conservation. Colour coding is shown in inset. Catalytic
triad residues, highly conserved surface-exposed residues, disulphide
bonds (and location of absent SS3 bond), and Y200 residue are shown
as sticks. Left and right views are orthogonal. Views looking at the active
site show molecules in similar orientations.