Biomedical Engineering Reference
In-Depth Information
A
B
P1
P1
C
P1
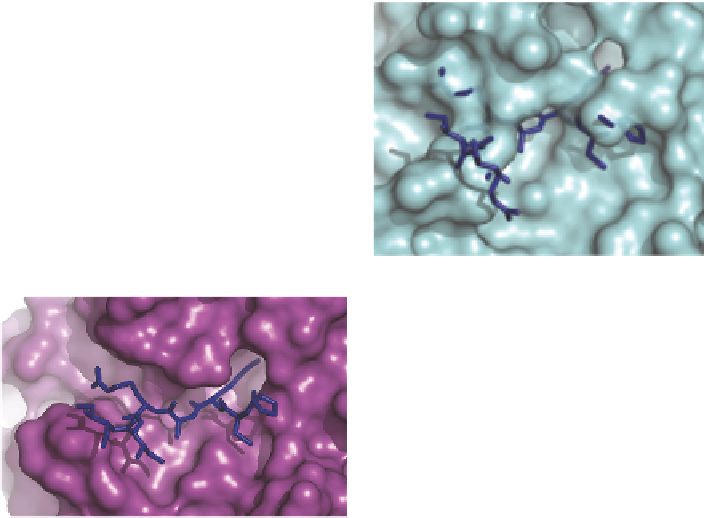
Figure 10.2
Blockage of SMIPP-protease inhibitor binding. Surface representations
of (A) SMIPP-S-I1 and (B) SMIPP-S-D1, showing modelled peptide
(sticks) and protein (cartoon) trypsin inhibitors. (C) Interactions between
trypsin and inhibitor (2PTC) shown for contrast.
10.3 Peptide Phage Display Screening Does Not
Identify Binding Partners
Phage displayed peptide libraries have been used previously to identify peptide
substrates of several chymotrypsin-like serine proteases.
14,15
Since it was likely
that the SMIPPs were inactive due to mutations to their catalytic residues, it is
important to investigate whether 20mer peptides fused to the minor coat pro-
tein pIII in a phage display library
16,17
could bind to the SMIPPs. Screening of
the phage display peptide library with three different SMIPPs did not
demonstrate a preference for any of the amino acid sequences displayed.
During successive rounds of panning, titrations showed that the pfu/ml counts
for a positive control known to select phage displayed peptides from the library
(5G8
16
) increased very quickly and remained high, while the pfu/ml of phage
displaying peptides that bound to the SMIPP-S proteins did not increase. While
phage displayed peptides that have an anity for binding to 5G8 increased in
number during each round of panning, the same was not found for the
SMIPPs. These results are consistent with the structural data described above,
implying strongly that SMIPPs do not bind peptides, as might be expected for a
catalytically inactive protease with an otherwise fully formed active site.