Biomedical Engineering Reference
In-Depth Information
O
OH
O
?
O
1
+
O
O
2
eozymes
His
Ser
Asp
Tyr
H
2
O
Lys
Lys
Active site
ensemble
3
Scaffold
ensemble and
matching
4
5
De novo
artificial
enzymes
6
TIM barrel
Jelly roll
Experimental
testing
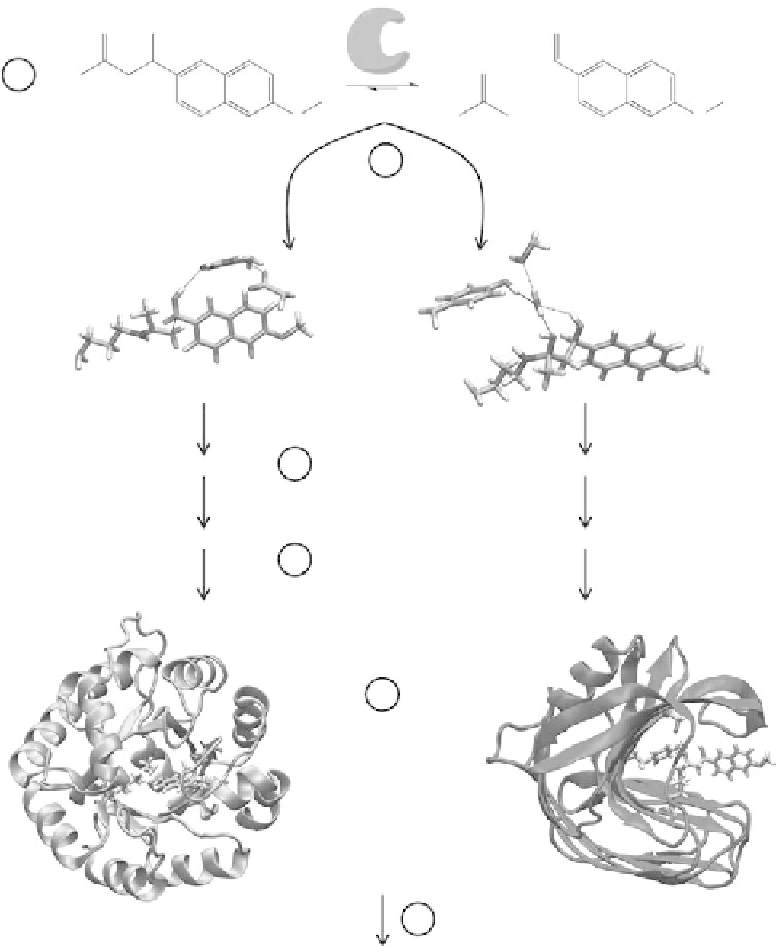
Figure 3.4
(See companion CD for color igure.)
. Overview. of. de. novo. computational. enzyme. design. protocol. for. the.
Rosetta. enzymes.. The. irst. step. (1). is. to. choose. a. reaction. that. the. new. enzyme. will. catalyze,. and. identify.
the.transition.state(s).and.key.intermediate(s).of.the.reaction.pathway..Possible.functional.groups.that.might.
stabilize. the. transition. state. are. identiied. by. chemical. intuition.. QM. calculations. (2). are. used. to. guide. and.
optimize.the.positioning.of.different.side.chains.and.functional.groups.around.the.transition.state,.generating.
different.possible.theozymes..Next,.an.ensemble.of.active.sites.(3).is.created.by.varying.the.side.chain.rotamers,.
and.these.active.sites.are.matched.to.complementary.protein.scaffolds.(4)..The.resulting.promising.models.are.
identiied.(5).and.tested.experimentally.(6)..The.coordinates.for.the.theozymes.and.the.enzyme.models.were.
kindly.provided.by.David.Baker.