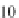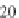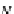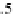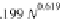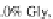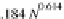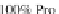Biomedical Engineering Reference
In-Depth Information
Fig. 1.3
Simulation results
forchainsrichinProlineand
Glycine residues
is comparable to the 3.0 Å rise per Proline from atomistic simulations and 3.1 Å
per Proline from experimental data (Dolghih et al.,
2009
). The dependence of the
radius of gyration of poly-Proline proteins on the sequence length is also studied in
Fig.
1.3
. It clearly shows the higher dimensions of these synthetic proteins compared
to natural proteins. It should be noticed that since Proline is considered a hydropho-
bic amino acid and the influence of solvent is not included in the present model,
the predicted gyration radius is overestimating the
R
g
of poly-Proline proteins in
aqueous solution.
1.4 Conclusion
In this paper, we have presented an implicit solvent, one-bead per amino-acid
coarse-grained model to study the unfolded state of proteins. To ensure that the
CG bending and torsion potential functions for bonded interactions are not biased
to any specific secondary structure, the obtained potentials were extracted from Ra-
machandran data of the coil regions of proteins. The potential functions have been
developed by accounting for the effect of neighboring residues, rendering the model
to be residue- and sequence-specific. The model has been used to study the correla-
tion between sequence composition and dimension of denatured proteins. Based on
the Proline and Glycine content of the protein sequence, an upper and lower bound
is constructed for the ensemble average
R
g
of denatured proteins, which is in agree-
ment with the available experimental data. The developed model sets the stage for
further developments towards the inclusion of electrostatic and hydrophobic inter-
actions to study the characteristics of natively unfolded proteins under physiological
conditions.
References
Adzhubei AA, Sternberg MJ (1993) Left-handed polyproline II helices commonly occur in globu-
lar proteins. J Mol Biol 229:472-493
Avbelj F, Grdadolnik SG, Grdadolnik J, Baldwi RL (2006) Intrinsic backbone preferences are fully
present in blocked amino acids. Proc Natl Acad Sci USA 103:1272-1277