Biomedical Engineering Reference
In-Depth Information
suspicion that there is something that is not quite right with this story. At the very least,
it seems to be too good to be true. In away, you are right. It is too good to be completely
true. Let me now try to pull the rug out from under the stack of cards that I have built.
Let us go all the way back to our original example where we had cell age versus
DNA and cell age versus cell number. I showed how we can remap the cell age to a
probability state axis. The boundaries of these states were originally defined by cell
age, but later on, I showed that we can compute these boundaries without using cell
age. That was the point where I left something very important out of the story. It is now
time for a full disclosure.
14.5.3 Loss of State Sequence Information
At the moment we make our measurements, we lose time or, for our example, cell
age. What this means is that there is no information in the data that allows us to
sequence our states. When we lose time, it is like pulling the string out from the
necklace of states that define our model. Equivalent data can be synthesized by any
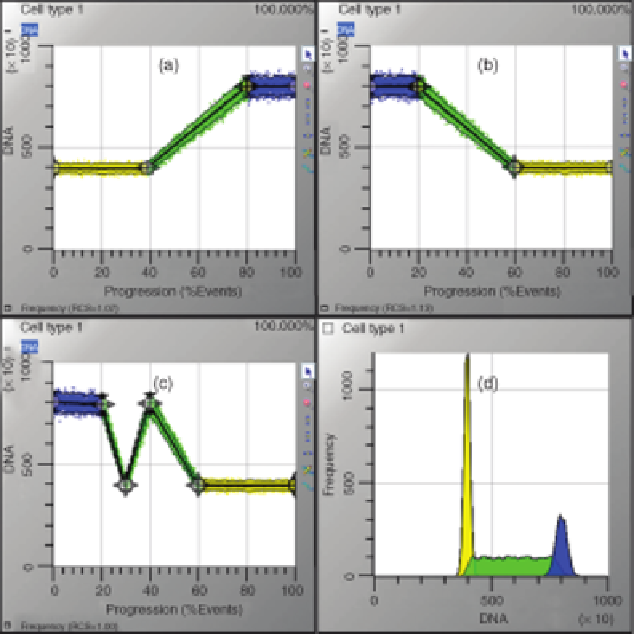
random sequence of these states. Figure 14.8 shows three examples of different state
FIGURE 14.8
Different models can produce equivalent data. The three models (a, b, and c)
can produce equivalent data (d) because listmode data no longer contain state sequence
information. (See the color version of the figure in the Color Plates section.)
Search WWH ::

Custom Search