Biomedical Engineering Reference
In-Depth Information
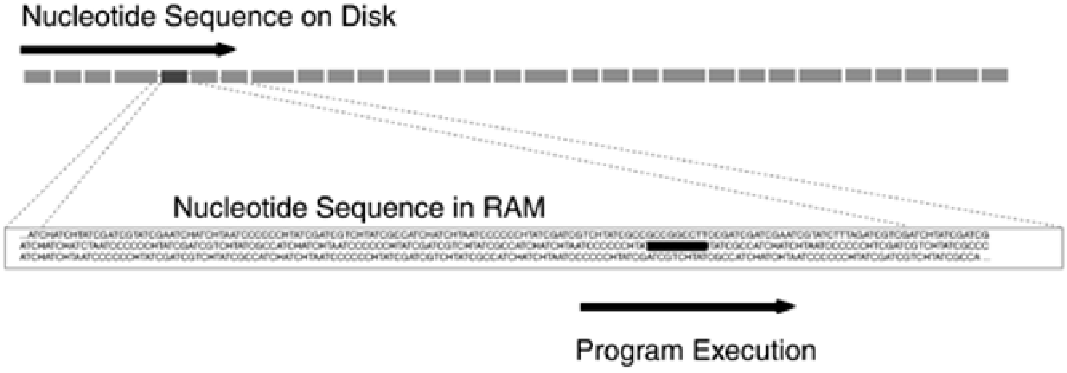
used for program execution, whereas an expansive disk or other non-
volatile memory serves as a container for data that can't fit in working
memory.
Volatility, working memory, and the volume of data that can be handled are key variables in memory
systems such as databases. In addition, there is the quality of interrelatedness; just as the genes in
the chromosomes are associated with each other by virtue of their physical proximity, the data in a
database are interrelated in a way that facilitates use for specific applications. For example,
nucleotide sequences that will be used in pattern-matching operations in the online sequence
databases will be formatted according to the same standard—such as the FASTA standard.
As reflected in the data life-cycle model discussed earlier, the data-archiving process involves
indexing, selecting the appropriate software to manage the archive, and type of media as a function
of frequency of use and expected useful life span of the data. From an implementation perspective,
the key issues in selecting one particular archiving technology over another depends on the size of
the archive, the types of data and data sources to be archived, the intended use, and any existing or
legacy archiving systems involved. For example, the size of the archive is measured in terms of the
number of items and the space requirements per item. Text-only archives of nucleotide or amino acid
sequences generally require less space per item than archives of 3D images of protein molecules and
other multimedia. Not only are space requirements generally much greater for multimedia data than
they are for text, but images usually require additional keywords and text associated with them so
that they can be readily located in an archive.
Similarly, a single source of data is generally much easier to work with than data from multiple,
disparate sources in different and often non-compatible formats. In addition, hardware and software
used in the archiving process should reflect the intended use of the data. For example, seldom-used
data can be archived using a much less powerful system, compared to data that must be accessed
frequently. Finally, it's rare to have the opportunity to initiate a digital archiving program from
scratch. Normally, there is some form of existing (legacy) system in place whose data has to be
converted to be suitable for archiving.
The simplest approach to managing bioinformatics data in a small laboratory is to establish a file
server that is regularly backed up to a secure archive. To use the hardware most effectively,
everyone connected to the server copies their files from their local hard drive to specific areas on the
server's hard drive on a daily basis. The data on the server are in turn archived to magnetic tape or
other high-capacity media by someone assigned to the task. In this way, researchers can copy the
file from the server to their local hard drive as needed. Similarly, if the server hardware fails for some
reason, then the archive can be used to reconstitute the data on a second server.
As noted earlier, from a database perspective, file servers used as archives have several limitations.
For example, because the data may be created using different applications, perhaps using different
formats and operating systems, searching through the data may be difficult, especially from a single
interface other than with the search function that is part of the computer's operating system. Even