Biomedical Engineering Reference
In-Depth Information
biotech enterprise composed of variety of commercial and in-house applications, a given data
element may be defined differently within different applications.
Patient age might be defined in months within a clinical pathology system, whereas patient age
within the microarray database and the data dictionary might be represented in years. The data
dictionary can be used to reconcile the two systems, providing an appropriate data transform
between the two representations. For example, the appropriate transforms to move between the
representations used by the pathology and microarray systems for patient age might be:
PatientAge (Data Dictionary) = PatientAge (Microarray) = PatientAge (Pathology)/12
The data dictionary can also impose a standard vocabulary on the system so that clinical findings can
be identified unambiguously. For example, one clinical system might refer to heart attack as "M.I.,"
another as "Myocardial Infarction," and yet another as "Heart Attack." By imposing a standard
vocabulary, the data dictionary allows data from the various systems to be combined into a unified
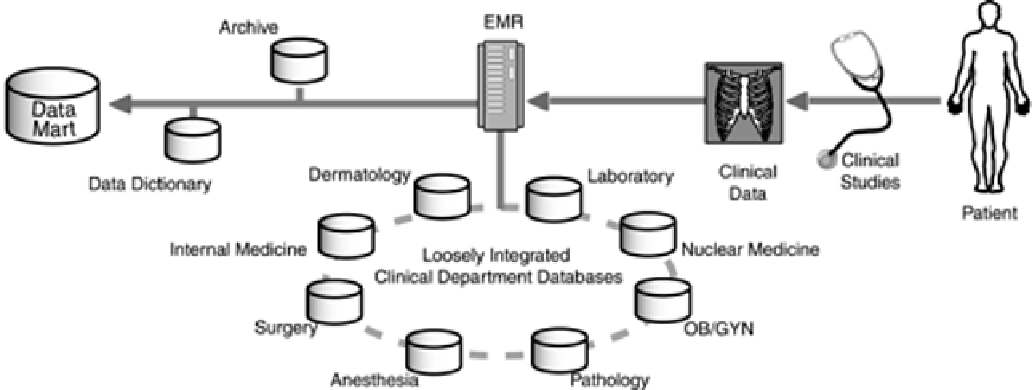
view of the patient that can be more easily mined for patterns. This view is typically maintained in a
data mart, as illustrated in
Figure 2-4
. The data mart contains a subset of the data that resides in the
individual databases combined with contents from these databases translated into a standard format
that can be efficiently mined for data.
Figure 2-4. Integration of Clinical Data. To create an EMR capable of
supporting efficient data mining, a data dictionary is used to impose a
standard format and vocabulary on data stored in the clinical data mart.
A parallel situation exists in the bioinformatics component of the patient data management. As
depicted in
Figure 2-5
, patients provide DNA source material for analysis in the form of tissue
samples, which are processed for microarray analysis, generating thousands of data points. These
data are then processed by a pattern-recognizer program to identify significant patterns. Researchers
rely on local databases of gene expression, medical relevance, and a data dictionary to provide a
common language and format for the data. Links to the large public genomic databases provide
additional reference material. As with the clinical data, the composite genomic data are stored in a
data mart for efficient manipulation and analysis through a suite of applications. Ideally, relevant
data from clinical applications are combined in the data mart as well.
Figure 2-5. Integration of Bioinformatics Data. Like clinical data,
bioinformatics data from a variety of sources and in numerous formats are
combined in a data mart to enhance data management.