Biomedical Engineering Reference
In-Depth Information
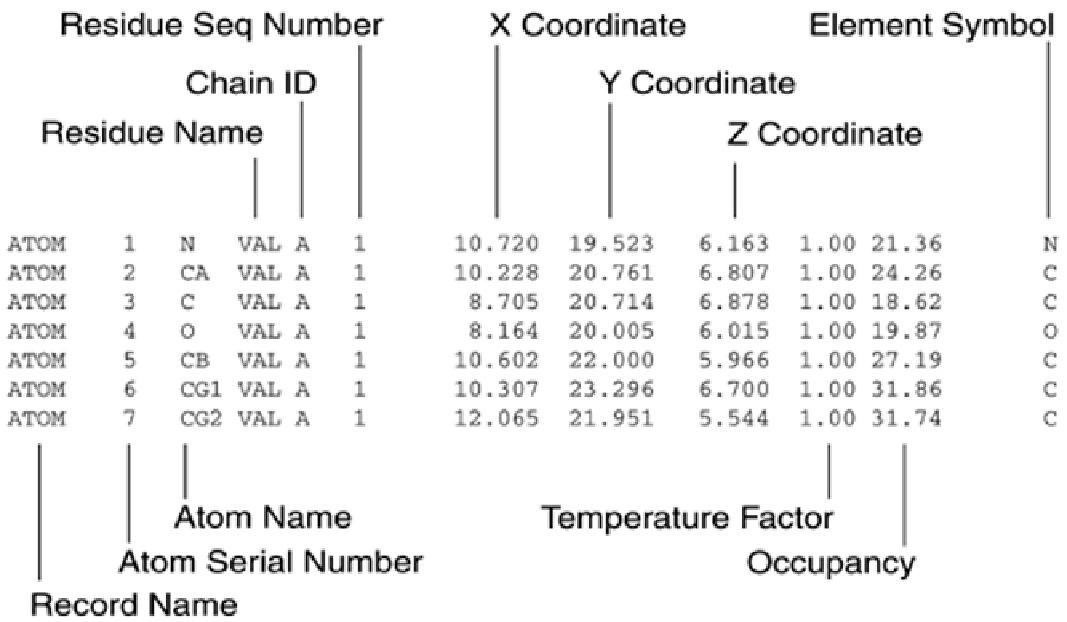
Whereas the Cartesian coordinate system maps atoms involved in a protein structure relative to an
absolute special geometry, internal coordinate schemes define all angles and positions relative to an
arbitrary structure, such as the first bond in an amino acid sequence. The use of an internal
coordinate scheme often provides a computational advantage over a Cartesian system, especially
when thousands of atoms are involved. However, working with an internal coordinate system often
makes it difficult to relate protein structures that are not connected, and it's difficult to determine
absolute distances between molecules and atoms.
The assumption that a protein's secondary structure can be completely defined as a function of bond
lengths, bond angles, and torsion angles, while not always valid, greatly simplifies the computations
involved. However, in some instances, even limiting consideration of protein structure to bond
lengths, bond angles, and torsion angles is too computationally intensive. For example, modeling
protein-protein interactions, with each protein molecule composed of perhaps several thousand
atoms, in an aqueous environment with several hundred-thousand water molecules, is currently
practically impossible on desktop hardware and may require days of supercomputer time. As a means
of simplifying the computations, protein molecules are commonly simplified by representing certain
chemical groups as points or ellipses that are either attracted to or repelled by surrounding water
molecules.
The overall process of determining or predicting tertiary protein structure from a known primary
structure or sequence is illustrated in
Figure 9-16
. Given a sequence of amino acids, the first step is
to generate a reasonable secondary structure by using bond lengths, angles, and torsion angles. The
next phase of the process, generating the tertiary structure, involves methods such as molecular
dynamics and Monte Carlo methods to create a library of tertiary protein structure candidates.
Figure 9-16. General Ab Initio Protein Structure Prediction Process.