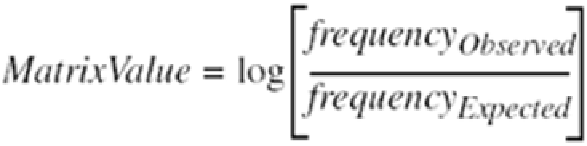Biomedical Engineering Reference
In-Depth Information
V
Valine
W
Tryptophan
X
Unknown
Y
Tyrosine
Z
Glutamate or Glutamine
The figures used to populate the matrices are based on the formula:
A matrix value of "0" signifies that a substitution typically occurs at a random base rate, whereas a
negative matrix value infers that the substitution is less likely than by chance alone. A positive matrix
value means that the substitution occurs more often than suggested by chance. For example, in the
PAM-250 matrix, A and N (Alanine and Asparagine) substitute for each other at a rate (0) that is
expected by chance alone. Conversely, A and W (Alanine and Tryptophan) substitute for each other
at a rate (-6) much lower than expected for a random substitution.
From the perspective of supporting sequence alignment, the main diagonal reveals the relative value
of maintaining matches in the pairwise sequence alignment process. For example, in the PAM-250
substitution matrix, given a choice of shifting a sequence by adding gaps or other means that affect
either an A-A (Alanine-Alanine) alignment or a C-C (Cysteine-Cysteine) alignment, the C-C alignment
should not be disturbed. This is because the C-C alignment is rated at 12, compared to only 2 for the
A-A alignment.
Figure 8-5 The Percent Accepted Mutation Substitution Matrix 250 (PAM-
250).
PAM-250 SUBSTITUTION MATRIX
A R N D C Q E G H I L K M F P S T W Y V B Z X
A 2
R -2 6
N 0 0 2
D 0 -1 2 4
C -2 -4 -4 -5 12
Q 0 1 1 2 -5 4
E 0 -1 1 3 -5 2 4
G 1 -3 0 1 -3 -1 0 5
H -1 2 2 1 -3 3 1 -2 6