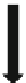Hardware Reference
In-Depth Information
Fig. 3.5
Modules and their
interconnections for the
dictionary-based error
recovery system
Movements of droplets
Sensor on the biochip
Droplet checking
results
FSM
Address of the dictionary entry
needs to be accessed
De-compaction module
…
Dictionary entry for
error recovery
Actuation signals for
the biochip
3.6.1
Sensing Module
The sensing module can be designed on a biochip based on different sensing
techniques. Here, we use the imaging-based droplet detection method as the
example to illustrate the implementation of sensing modules in FPGA. Note that
the error-recovery algorithm approach proposed in this chapter is general and it can
be applied to cyberphysical biochips with various sensing systems.
In Sect.
2.2.1
, an imaging-based droplet-tracking algorithm is discussed. During
the execution of the bioassay, an image sensor is used to monitor the entire biochip.
Based on the images captured by the image sensor, the control software can
automatically search for droplets by “template matching” (Sect.
2.2.1
). The acquired
images are first converted to two-dimensional matrices. For example, an image with
M
w
N
w
pixels will be converted to an M
w
N
w
matrix, in which each element
represents one pixel of the image.
An image of a typical droplet on the biochip is selected as the “template” (which
is written as T ). Each time, the software crops a sub-image (T
s
) from the image of
the entire biochip. The cropped sub-image and the template image are considered as
two vectors, and the correlation index of these two vectors represent the similarity
between the template and the cropped sub-image. In order to locate the positions
of droplets on the biochip precisely, the correlation index is calculated on a pixel-
by-pixel basis [
14
]. Therefore, if the image for the entire biochip has M
w
N
w
pixels and the template image has M
s
N
s
pixels, the calculation of the correlation
factor between the template image and the cropped sub-image will be implemented
.M
w
M
s
/
.N
w
N
s
/ times. The computational complexity of droplet tracking is
significantly high when there is a large number of pixels in the image of the entire
biochip, hence the image-based droplet-tracking method need to be performed by
the software on a computer [
14
].
Based on the characteristics of the digital microfluidic biochip, the image-
based droplet-tracking procedure can be simplified and accelerated. Hence all the
calculations can be performed on the FPGA. The movements of droplets on the