Biology Reference
In-Depth Information
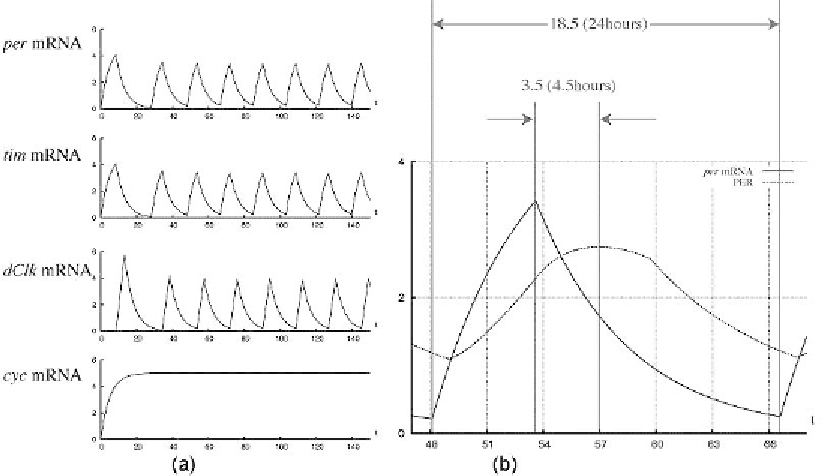
Fig. 7. (a) Behaviors of concentrations of four mRNAs obtained from simulation by GON. (b) By reference to scale markings
of time, time difference around four and half hours is observed between the peaks of concentrations of
per
mRNA and PER.
Complex forming rate of dCLK/CYC at transition
T
1
is realized by the function
m
2*
m
4/20, where
m
2 and
m
4 are amounts in places dCLK and CYC, respectively. Complex forming rates of PER/TIM
and PER/DBT at the transitions
T
2
and
T
3
are realized similarly. Transitions
T
4
,
T
5
, and
T
6
represent
the degradation rates of complexes of the corresponding proteins. Figure 7a is the simulation result
of the HFPN in Fig. 6. It indicates that this HFPN model representing two negative feedback loops,
the PER-TIM feedback and the dCLK-CYC feedback, successfully produce periodic oscillations of
per
mRNA (
m
6),
tim
mRNA (
m
8), and
dClk
mRNA (
m
1), while the concentration of
cyc
mRNA (
m
3)
keeps constant expression.
It is known that the protein TIM stabilizes phosphorylated PER by dimerizing with it. This phenomenon
is reflected to the firing speed of transition
T
5
, that is, the firing speed of transition
T
5
(
m
13/15) is set
to be slower than the one of transition
T
7
(
m
7/10). Moreover, it is suggested in [13] that the normal
function of protein DBT is to reduce the stability and thus the level of accumulation of monomeric PER
proteins. This function is realized in Fig. 6 in transition
T
3
. It is clearly expressed in Fig. 7b that there
is time difference around four and half hours between the peaks of concentrations of
per
mRNA and
PER which is believed to be arisen from the two facts mentioned above. This indicates that the result of
simulation is in good agreement with the experimental observation reviewed in [11].
Price
et al.
[13] discussed properties of
dbt
L
and
dbt
S
which are mutants of the gene
dbt
and showed
that transcription of the gene
per
is affected by these mutants. That is, period of
per
mRNA in
dbt
L
mutant (
dbt
S
mutant) is longer (shorter) than the one in wild type. The behavior of
per
mRNAs in these
two mutants and wild type is described in Fig. 8. It is obtained by changing the formula at transition
T
3
.
These simulation results suggest that circadian rhythm is controlled by the complex forming rate of PER
and DBT proteins, which is affected by the mutants
dbt
L
and
dbt
S
.