Biology Reference
In-Depth Information
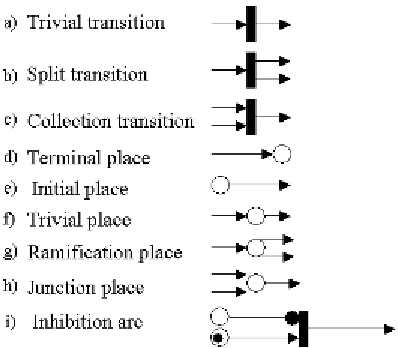
Fig. 1. Components of a Petri net. Their counterparts in metabolic networks are as follows: a) A
→
B (isomerization); b) A
→
B
+
C; c) A
+
B
→
C; d) a product that is not further consumed; e) a substrate that is not produced; f) a metabolite produced
and then consumed; g) a metabolite produced in one reaction and then consumed in two or
n
reactions; h) the situation opposite
to g); i) inhibition phenomena.
A simpler solution is offered by Petri net theory [Reisig, 1985; Starke, 1990]. Two kinds of nodes
are considered: places and transitions. The nodes and arcs between them represent the static structure,
while some more elements/components such as tokens indicating time-dependent weights of places are
used to describe the dynamics. Beside place/transition nets, also condition/event Petri nets have been
proposed in the literature. Here, we only deal with the first type. Petri nets can be employed for the
graphical description of processes. They allow us to understand more intuitively the temporal evolution
of systems by considering flows of tokens through the nets. They offer also an appropriate formalism
for the analysis of biochemical networks, as has been pointed out earlier by several authors [Genrich
et
al.
, 2001; Heiner
et al.
, 2000; Heiner
et al.
, 2001; Hofest adt, 1994; K uffner
et al.
, 2000; Oliveira
et al.
,
2003; Peleg
et al.
, 2002; Reddy
et al.
, 1996], while the above-mentioned modelling approaches [Fell
and Wagner, 2000; Heinrich and Schuster, 1996; Jeong
et al.
, 2000; Leiser and Blum, 1987; Schuster
et al.
, 2002a, 2002b; Teusink, 1998] are independent of Petri net theory. In the present paper, we shall
show the correspondence between concepts in both, Petri nets theory and traditional metabolic network
analysis. Some examples will help the reader see the similarities and to exploit them.
In the present paper, we shall focus on the use of Petri nets for the topological analysis of biochemical
networks rather than for the analysis of the dynamic behaviour. In particular, we shall deal with various
invariants and other features in these nets such as boundedness and liveness and reveal their biochemical
meaning. Moreover, we shall discuss the appropriate treatment of source and sink metabolites.
SIMILARITIES BETWEEN PETRI NET THEORY AND TRADITIONAL BIOCHEMICAL
MODELLING
In graphical representations of Petri nets, circles are used for places, while rectangles stand for
transitions (Fig. 1). The correspondence place - substance (in biochemistry often called metabolite) and
transition - reaction/enzyme is obvious. Metabolic networks have a static level - the stoichiometry, and
a dynamic one, characterized by fluxes. The stoichiometric coefficients indicate how many molecules