Biology Reference
In-Depth Information
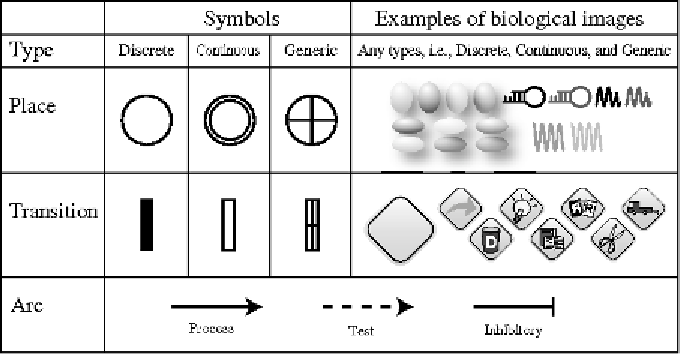
Fig. 1. In Cell Illustrator Petri nets are constructed using three kinds of symbols for entities, processes, and connectors. Discrete,
continuous, and generic types are associated to entities and processes, and direct, associate and inhibitory types are associated
to connectors. Additionally, entities and processes can be replaced with pictures of biological ontology terms from the Cell
System Ontology [Jeong
et al.
, 2007]. This information enhances both the human readability and machine readability of the
HFPNe biological pathway models built in Cell Illustrator.
Arcs are directed connections from places to transitions (input) and from transitions to places (output).
Places can contain tokens, and a transition which has all places connected by input arcs with tokens, will
transfer these tokens (in discrete units) to the places connected by output arcs.
Since the proposal of the original Petri net [Petri, 1962], various types of Petri nets have been developed,
e.g. timed Petri net, continuous Petri net and Hybrid Petri net (HPN) [Nagasaki
et al.
, 2005]. Hybrid
Petri nets have new types of place and transition that can receive continuous token values, along with
the classic discrete ones. Like other proposed Petri nets, Hybrid Petri nets also expand the original Petri
net by introducing the concepts of “inhibitory arc” and “test arc”; the inhibitory arc will stop a transition
from working if the value on the place it is connected to (its input) is higher than a certain threshold
(weight in Petri net terms), which in biology might easily represent a transcriptional repressor or an
enzyme inhibitor. The test arc will not consume any tokens from the place it is connected to, making an
easy analogy to an enzyme, which is not consumed by the reaction it catalyses.
Pioneering works by the groups of Reddy and Hofest adt were among the first to apply Petri nets to the
modelling of biological pathways [Reddy
et al.
, 1993; Hofest adt and Thelen, 1998]. An advanced HPN
has been also applied to the modelling of lambda phage pathway [Matsuno
et al.
, 2000]. Furthermore,
Hybrid Functional Petri nets (HFPN) and its extension (HFPNe) were developed by expanding the HPN
to be more powerful and suitable for biological pathway modelling and simulation [Matsuno
et al.
, 2003;
Nagasaki
et al.
, 2004].
HFPN cannot only handle both discrete and continuous events at once, but also allows any kind of
functions to be assigned to the delay, weight and speed parameters. HFPNe was introduced to facilitate
the handling of any kind of objects within the concept of a Petri net. To handle these objects, new
generic elements for place and transition were introduced (see Fig. 1). Using HFPNe, complicated
biological pathway processes can be modelled, for example, networks involving gene regulation, signal
transduction and metabolical reactions, as well as other biological processes that are not normally treated
in biological pathways, such as alternative splicing and frame-shifting [Nagasaki
et al.
, 2004].
Differential equations can be easily modelled using a subset of HFPNe elements, by assigning contin-
uous values for place and transition and suspending the weight parameters evaluation. Nagasaki
et al.
,