Biology Reference
In-Depth Information
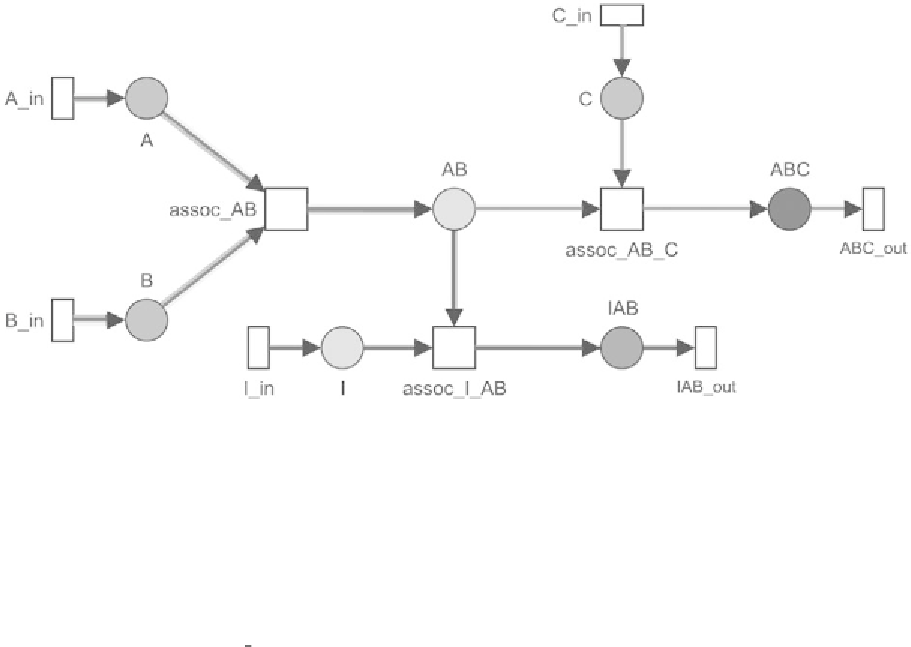
Fig. 4. Reduced PN module which describes the formation of an inhibiting intermediate complex (
IAB
). The blue highlighted
pathway covers reactions, which result in a functional target complex
ABC
, while the red colored pathway describes a T-invariant
covered by reactions, which result in a non-functional complex
IAB
. (Colours are visible in the online version of the article at
(b)
Allosteric enhancement
describes the presence of a specific protein factor, which increases
the affinity of other proteins to participate in subsequent reactions (
e.g.
, subcomplex formation,
RNA recognition etc.). The structural analysis gives four T-invariants, two of which produce
the target complex
AB
. The interaction of factors
A
and
B
can result in a dimerized complex
AB
(named “
assoc AB
”, Fig. 5a). However, the enhancer may be necessary for the protein
(complex) to be active. The model accounts for the presence of enhancer
E
with a higher output
of
AB
(Fig. 5a, blue pathway) due to an increased arc weight. Hence, transition
AB out
has to
fire twice to reproduce the initial marking. Biologically, this can be interpreted as an increased
signal transduction as
AB
reaches a state of higher disposition for participating in downstream
reactions. The reduction of the system for the dissociation reaction of the dimer
AB
decreases
the number of T-invariants by one (Fig. 5b). Two T-invariants involve transition
assoc AB
,but
only one produces
AB
, while the other purges
AB
from the network (Fig. 5b, red pathway). The
pathway involving
E
via reaction
assoc ABE
produces an increased amount of
AB
. Thus, the
reduced model captures all essential aspects of the enhancer dependent complex formation.
2.
Enzymatic reactions
describe the reactions, in which a catalytically active enzyme acts on molec-
ular groups of spliceosomal proteins,
e.g.
, kinases phosphorylate proteins. This behavior was
modeled as loop, which preserves the marking of the respective place and results in a simple
conservation relation (Fig. 6a). Also, helicase-like proteins with DExD/H box domains have been
frequently found in purified spliceosomes [Jurica and Moore, 2003], and were considered as sep-
arate module. This module was extended by another reaction, representing the enhancement of
substrate specificity of the helicase (see Fig. 6b). In this context, the rate of NTP hydrolyzation
has been proposed as a crucial parameter for splicing fidelity, since fast kinetics on weak or in-
correct protein-substrate interactions increases the chance of dissociation of essential spliceosomal
proteins. In consequence, such defective substrates could be submitted into a degradation pathway
[Staley and Guthrie, 1998]. While a putative degradation pathway was integrated as a branch into