Biology Reference
In-Depth Information
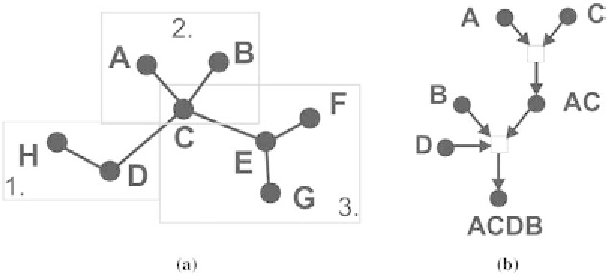
Fig. 1. Two types of protein interaction networks: (a) An undirected network of binary interactions, e.g., modeled in [Zhang
et
al.
, 2007]. (b) A directed hierarchical molecular interaction network represented as Petri net.
this context, the term “transcription factory model” has been coined, denoting the concerted action of
transcription and splicing machinery at the places of active gene transcription [Bentley, 2002]. The
connection and coupling of different protein machines in the cell emphasizes the complex environment
in which spliceosomal assembly is embedded.
The vast amount of experimental data makes it necessary to structure the information for deriving
new hypotheses about the underlying signal transduction processes. This task requires the development
of theoretical models, which are able to integrate much of the existing data while being suited for
rigorous validation and stepwise extension. Modeling goals can be summarized in
i
) visualization,
ii
)
comprehensible data annotation
iii
) data abstraction, and
iv
) model simulation, allowing mathematical
description of the model. In particular, alternative splicing often involves different sets of splicing factors
in addition to the spliceosomal core components. Hence, variations of the network should provide a
sound base for testing new hypotheses on regulation of splicing and AS events.
Several structural and kinetic factors were proposed to influence splicing patterns of a gene such as
i) precise balance and concentrations of regulatory proteins ii) the nature of interaction like inhibiting
or cooperative effects iii) the number of interactions, which define the connectivity of a network iv) the
speed of transcriptional elongation or recruitment of splicing factors [Park
et al.
, 2004; House and Lynch,
2007]. Additionally, a temporal component can be accounted as the assembly pathway bears a number of
timed dependencies. These key points pose an essential base for modeling spliceosomal processes, but
most of them can presently not be comprehensively be applied. Incompleteness of experimental data, for
example, the lack of knowledge about interaction kinetics, concentrations or the exact temporal order of
reactions is accompanied by heterogeneity of proposed mechanisms for different stages of spliceosome
assembly.
Protein interaction databases such as S
TRING
[von Mering
et al.
, 2007], cross-referencing to M
INT
,
H
PRD
,B
IO
G
RID
,D
IP AND
R
EACTOME
,A
PID
[Prieto and Rivas, 2006] or I
NT
A
CT
[Kerrien
et al.
, 2007], al-
ready provide an extensive organisation and integration of experimental and predicted protein interaction
data. However, most of them represent static interactions (Fig. 1a) without providing information about
the temporal order and direction of interactions within hierarchical networks. This underscores the need
of finding ways to incorporate additional knowledge from literature, what in turn helps in the analysis
of hierarchical processes by providing insights into the progression of signals and protein interactions in
networks such as spliceosome assembly.
Figure 1a depicts an unordered-undirected protein-protein interaction network as reviewed in [Zhang
et al.
, 2007], and a directed ordered interaction network presented in this work (Fig. 1b). Due to lack