Biology Reference
In-Depth Information
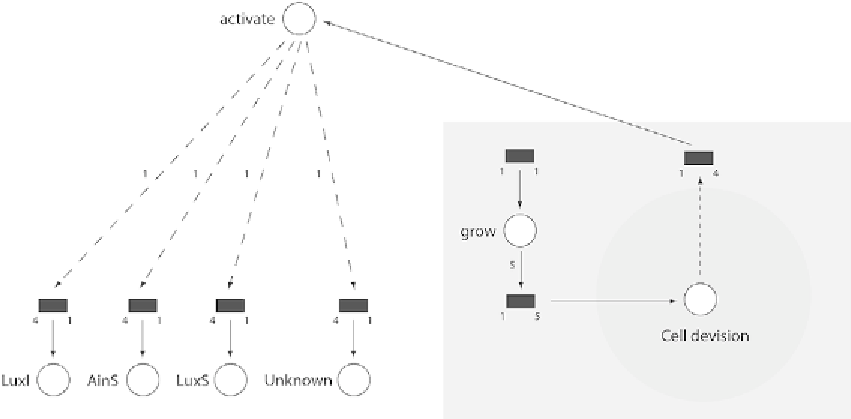
Fig. 14. A one-dimensional array of sub Petri nets for including growth and cell division in the model*.
Based on the work presented in this paper, the use of the Petri net models for the analysis of cell-
to-cell communication processes was demonstrated. Besides the traditional modeling and simulation
approach based on ordinary differential equations (ODEs), the approach of using Petri nets and logic
based descriptions becomes more and more important. For cell-to-cell-communication processes, where
the lack of laboratory data is a common problem, it is particularly difficult to formulate a sophisticated
mathematical system.
We showed that the cell-to-cell communication process and information flow within a cell and across
cell colonies can be modeled and simulated with Petri net network structures. The models are examples
of how the behavior of cells modeled by a Petri net can be simulated. Ideas for laboratory experiments can
be gained and tested by changing the basic Petri net. One approach is a theoretical knockout experiment.
For this task knocked out genes are modeled by deleting the corresponding transitions. The changed Petri
nets can again be combined as mentioned above and the simulation results can be compared. A change
of gene sequences influencing the catalytic efficiency can be modeled by changing speed parameters of
the corresponding transition. Factors outside the cells that influence the signal molecules, the diffusion
speed of the medium, the availability of nutrients or even processes inside the cell can also be modeled.
In conclusion, the approach of using Petri nets for cell-to-cell-communication processes gives immense
possibilities for the future. It is a sophisticated modeling and simulation technique to represent signal
transduction, cellular rhythms and cell-to-cell communication.
ACKNOWLEDGEMENTS
The work is partially supported by the EU project “CardioWorkBench” (
http://www.cardioworkbench.
eu/
) and the BMBF Project (CHN 08/001). Ming Chen would like to thank the DAAD fellowship and
the NSFC for related project financial support.
*A colored version of the figure/chart is available at
In Silico Biol.
10
, 0003 <
http://www.bioinfo.de/isb/2010/10/0003/
>
, 1
Feb uary 2010.
r