Biology Reference
In-Depth Information
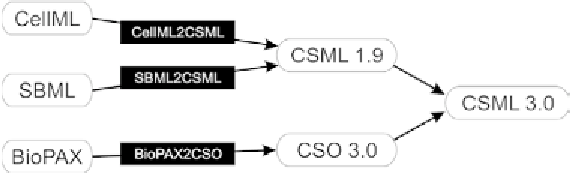
Fig. 5. XML format transformation. CellML and SBML are transformed with CellML2CSML and SBML2CSML to CSML
1.9 and BioPAX is transformed with BioPAX2CSO to CSML 3.0 without losing any information. Any models in CSML 1.9,
CSML 3.0, and CSO 3.0 run on CIO 4.0. All transformation tools such as CellML2CSML, SBML2CSML, and BioPAX are
included in CIO 4.0 and user can directly import any models to CIO 4.0.
TRANSPATH [31], can also be imported into CSML 3.0 format. It should be noted that these functions
in Cell Illustrator 4.0 have been employed and extended the results in several studies [32-35].
The
detailed internal conversion data flow of Cell Illustrator 4.0 is summarized in Fig. 5.
Cell Illustrator 4.0 is able to export a CSML 3.0 model into a CSO 3.0 model without loss of
information since CSML 3.0 is fully compatible with CSO 3.0. Other well-known XML formats, SBML
V3L3, CellML 1.1, and BioPAX L2, are not rich enough to hold the full CSML 3.0 model; thus,
exporting from CSML 3.0 to these formats results in loss of important information, which is not handled
in SBML and CellML. For example, none of these formats have unified graphical representation since
the model developed in one application cannot load on other application with the same view. Recently,
although SBGN L1 has been proposed [36], its format is still under development and is considered only
a limited graphical representation of the biological components. As another example, the BioPAX L2
cannot handle signal transduction reactions and also cannot deal with information related to pathway
simulation. From the user's point of view, more export functionality to other formats,
e.g.
, Cytoscape [37],
will provide better usability. This will be a task for a future release.
Cell System ontology and standard icons
The native XML format of Cell Illustrator 4.0 is CSML 3.0, which has the background of Cell System
Ontology (CSO) 3.0 [32]. CSO allows ontology based representation of signal transduction pathways,
gene regulatory networks, metabolic pathways, cell-cell interactions with kinetics, and graphical infor-
mation. Formally, the schema is defined using Web Ontology Language (OWL) [38]. The major features
of CSO are as follows:
1. CSO 3.0 can be applied to a pathway model both with and without simulation; and
2. the core vocabulary in a biopathway is prepared in CSO 3.0 as entities, processes, and cellular
locations (92, 275, and 114, respectively) and all terms in the vocabulary are equipped with
standard icons.
Feature (i) allows representation of a pathway model in the absence of kinetics,
e.g.
, KEGG [29] or
Reactome [39], without loss of information. Feature (ii) not only boosts more exact data exchange with
other applications (since the original feature of the ontology concept) but also enhances the intuitive data
exchange among users since standard icons completely remove the ambiguity of a graphical pathway
model,
e.g.
, the process icon always means phosphorylation among any applications that support CSO
3.0. In Cell Illustrator 4.0, all standard icons are collected in the Biological Element dialog (Fig. 6).
Thus, only by repeating drag and drop (D&D) operations of those icons from the dialog with a filtering