Biology Reference
In-Depth Information
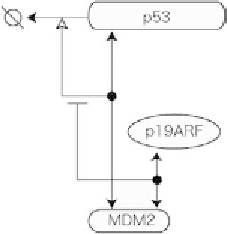
Fig. 6. Molecular interactions of proteins p53, MDM2, and p19ARF described by Kohn [Kohn, 1999] based on descriptions in
[Pomerantz
et al.
, 1998].
Pomerantz
et al.
[Pomerantz
et al.
, 1998] provided the result that protein p53 accumulates in a HeLa
cell after injecting gene
p19ARF
in the cell. In addition, they reported that protein p19ARF injection
increases protein p53 and the amount of products which are activated by protein p53. Figure 3(4) shows
an observation corresponding to that by Pomerantz
et al.
[Pomerantz
et al.
, 1998]. In this figure, the
complex p53-MDM2-p19ARF concentration keeps high level due to the complex formation between
p53, MDM2 and p19ARF, which allows protein p53 to escape from its ubiquitination by protein MDM2.
The increase of protein p53 (the part of p53-MDM2-p19ARF complex) coincides with the experimental
observation reported in [Pomerantz
et al.
, 1998]. However, for gene
Bax
, which is activated by protein
p53, the simulation result shows low expression while the gene expression of
Bax
is high in [Miyashita and
Reed, 1995]. Recall that we have assumed that the complex p53-MDM2-p19ARF has no transcriptional
activity for genes
Bax
and
MDM2
. This means that, in the HFPN simulation model, only protein p53 in
the nucleus (p53(N)) can activate
Bax
and
MDM2
. On the other hand, the simulation result of Fig. 3(5)
shows high expression of gene
Bax
as well as a certain amount of concentration of protein p53, being
consistent with the experimental observation in the literature [Pomerantz
et al.
, 1998]. Note that the
increase of p53-MDM2-p19ARF complex promotes not only the expression of gene
Bax
but also the
production of protein MDM2 in the nucleus. The increased MDM2 accelerates the decomposition of
p53 exported to the cytoplasm from the nucleus. Thereby a lower concentration of Fig. 3(5) than that of
Fig. 3(4) is induced. The simulation results suggested that protein p53 should have the transcriptional
activity in the forms of the trimer of proteins p53, MDM2, and p19ARF.
FURTHER REMARKS
Most existing pathway databases present many biological maps of molecular interactions. However,
in order to conduct simulations based on these maps, more biological facts such as reaction speeds of
complex formation and protein degradations have to be included to these maps. This means that we have
to reconstruct computational pathway maps for simulations after careful reading of papers of the interest.
In other words, these pathway databases have not been constructed on the assumption that pathways
included in them will be used for simulations.
The HFPN model of the complex p53-MDM2-p19ARF has been constructed based on biological
knowledge being extracted by careful reading of the literature. Of course, with no proof by biological
experiments, we could not conclude that the complex p53-MDM2-p19ARF has transcriptional activity
for genes
Bax
and
MDM2
. However, without the help of simulations, it is hard to get insights into the