Biomedical Engineering Reference
In-Depth Information
For proteins with the sequence length of 200-500 residues we have used RBF
network and the compositions of specific amino acid residues (Ala, Asp, His, Tyr,
and Val) for predicting the number of
-strands (10, 12, 14, 16, and 18). These five
amino acids have been selected based on the highest performance obtained with the
combination of amino acids using forward feature selection procedure. This method
could correctly identify the number of strands in 27 of the 28 considered proteins
and the leave-one-out cross-validation accuracy is 96%. In the second step, we have
used PSSM profiles and RBF networks for predicting the membrane spanning
segments. Our method could predict the membrane spanning residues with the
accuracy of 87%. The sensitivity and specificity are 82% and 90%, respectively.
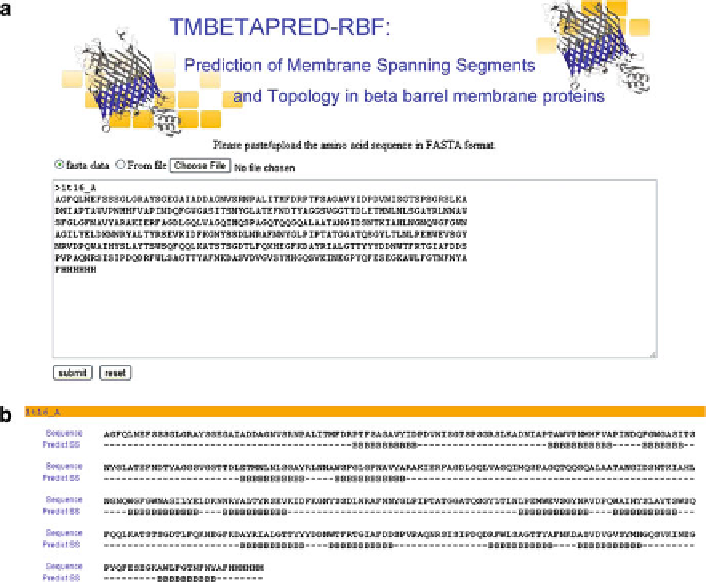
We have developed a web server for predicting the membrane spanning
b
-strands
for any
b
-barrel membrane protein and the details are presented in Fig.
4
.
It takes the amino acid sequence in one letter format as the input and automati-
cally omits gaps and numbers. The output provides the amino acid sequence of the
protein along with the predicted membrane spanning
b
-strand regions. The predic-
tion results are available at
http://rbf.bioinfo.tw/~sachen/tmrbf.html
.
The online resources available for predicting the membrane spanning segments
b
of
b
-barrel membrane proteins are included in Table
4
.
Fig. 4 Illustration for predicting the membrane spanning segments in
b
-barrel membrane
proteins. (a) Input sequence in FASTA format; (b) predicted
b
-strand segments