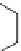Biomedical Engineering Reference
In-Depth Information
Roche et al. [
61
] employed different techniques such as self-organizing maps
(SOM), PCA, PLS, and supervised neural networks to develop predictive models
based on 1,258 descriptors to classify 472 compounds. The substructure analysis
performed with LeadScope did not highlight any moieties exclusive for blockers or
for nonblockers, although some weak trends could be noted. In 57% of the
nonblockers and in 30% of the blockers, two hydrogen-bond donors separated by
six bonds were present. In 20% of the nonblockers and in 2% of the blockers,
a benzenesulfonyl groups was found. In 49% of hERG inhibitors, the 1-R-4-alkyl-
benzene moiety was found. The best prediction performance was achieved with the
neural network model, which was able to correctly classify 93% of the nonblockers
and 71% of the blockers (95 compounds validation test set). The prediction method
was additionally used to analyze virtual combinatorial libraries to demonstrate its
applicability for shaping compounds libraries toward low probability to contain
potential hERG blockers. The structures based on scaffold one (Fig.
1
) have a
structural moiety common to many hERG inhibitors, while the molecules based on
scaffold two are designed to be nonblockers. The prediction highlights that in the
library of compounds based on scaffold one there are 58% of potential hERG
blockers, while in the second library the possibility to have hERG blockers
decreases to 0.1%.
Bains et al. [
59
] applied evolutionary programming with fragment-based
descriptors to predict hERG inhibition. The resulting model shows an accuracy of
85-90% for the classification of blockers and nonblockers. The model was
generated calculating 618 fragment- and nonfragment-based descriptors on 124
compounds randomly partitioned in 70:30:24 for training, generalization, and
validation data sets. Ten different partitions and subsequently 10 runs for each
partition were performed to generate a “consensus model” based on the average of
the prediction of the ten best generalizing models. The partition with the best
performance over the data sets was selected. Through a meta-SAR analysis, 30
descriptors related to hERG inhibition were identified. Inspection of the selected
descriptors revealed that the presence of a secondary or tertiary amino group, of one
or more aromatic rings and of a five-membered nitrogen heterocycle increases the
potency of hERG blockers. The model also shows that the presence of negatively
ionizable groups such as COOH and of oxygens as H-bond acceptors is detrimental
for hERG inhibition. Analysis of the 60 most correlated descriptors with hERG
blockade provided a pharmacophore model similar to the already published ones. It
consists of a nitrogen atom in the center to which an aromatic and a hydrophobic
feature are attached, separated by a linker of 4-5 and 1-2 carbon atoms, respec-
tively. This pharmacophore model highlights again the importance of the presence
of a secondary or tertiary amino group linked with two hydrophobic or aromatic
O
R2
N
N
N
Fig. 1 Chemical scaffolds
of the combinatorial libraries
R1
R2
R1
O
Scaffold 1
Scaffold 2