Biology Reference
In-Depth Information
that the majority of these QTLs are robust, being
identified in multiple populations from multiple
donor parents. Thus, the major
SKC1
QTL is seen
in nearly all populations derived from donors that
originate from India/Bangladesh, that is, Nona
Bokra, Pokkali, FL478 (derived from Pokkali),
Kala Rata, Chikiram Patnai, and Cheriviruppu
(Figure 3.1). The same is true for most QTLs;
they are typically shared between several donors
(Table 3.1). For example, regions around 11 Mb
(
Saltol
/
SKC1
) and 39 Mb on chromosome 1, 28
Mb on chromosome 2, 5 Mb and 30 Mb on chro-
mosome 3, 22 Mb on chromosome 6, and 8 and
25 Mb on chromosome 12 have been identified
from numerous populations. In some cases, there
is some doubt as to QTL position, as the mark-
ers used are difficult to accurately map to the
reference genomes or to other genetic maps. In
other cases, there may be more than one QTL
present in the interval (e.g., chr01L and chr03L).
Nevertheless, in the majority of these cases, the
same genes are probably responsible for these
overlapping QTLs.
The high incidence of overlap in QTL regions
probably reflects the shared origins of many
donors in India and Bangladesh, and their shared
physiological mechanisms (dominated by Na
+
exclusion). Those few studies involving donors
with no significant attachment to this geographic
region (such as IR64
×
Binam: Prasad et al. 2000; Zang et al. 2008) have
a much higher proportion of unique QTLs. In
×
Azucena and IR64
Fig. 3.1.
Comparison of QTLs on a) chromosome 1 and b) chromosome 3 identified in various mapping populations. For
a color version of this figure, please refer to the color plate.







































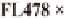










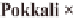

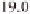








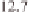
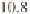








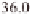

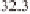

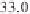

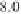











Search WWH ::

Custom Search