Agriculture Reference
In-Depth Information
LKP2
ZTL
StFKF1
StGI
?
StSP5G
?
StSP6A
StCDFs
StCOs
miR172
StBEL5
StPHYs
StRAP1
PotH1
Leaf
POTH1
StBEL5
StSP6A
Stolon
miR172
Key:
Repression
StGA20ox1
Induction
Interaction
Tr ansport
Tuberization
StGA3ox2
StGA2ox1
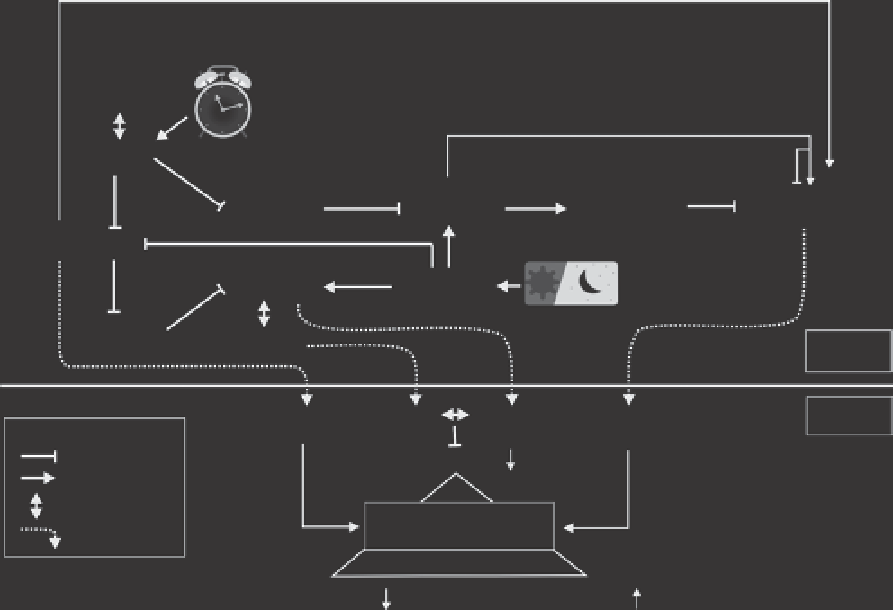
Fig. 4.3.
Schematic overview of identified genes associated with timing of tuberization and promoting
tuber formation. LKP1, ZTL and FKF1 are blue light receptors that are regulated by the clock genes. FKF1
and GI interact and bind to the transcription regulator CDF. The interaction of GI and FKF1 with CDF
targets this protein for degradation by the proteasome in the afternoon. During the early morning of long
days, CDF is not yet degraded and inhibits the expression of
CO
genes. CO protein is also a transcription
factor thought to be a specific inducer of the
FT
homolog,
StSP5G
. Evidence suggests that StSP5G
inhibits the expression of
StSP6A
, which in turn is considered the primary mobile tuberigen transported
to the stolon tip. Within the stolon tip, StSP6A activity is expected to trigger expression of tuber identity
genes through interaction with a yet unidentified partner.
StBEL5
,
POTH1
have also been shown to be
mobile and associated with transport to the stolon tip promoting tuber formation. PHYB plays a central
role in the regulation of a number of proteins inducing both
StCO
and
StBEL5
under the correct light
conditions (LD), while suppressing
miR172
in stolons under LDs, possibly through StRAP1 (Banerjee
et al.,
2006; Jung
et al.,
2007; Martin
et al.,
2009). At the stolon tip, hormones such as GA play a key role
in tuber initiation. It is known that the BEL-KNOX interaction is capable of suppressing GA production by
inhibiting
StGA20ox1
. The downregulation of
StGA3ox2
and the upregulation of
StGA2ox1
allows for
further rapid reduction of bioactive GA in the stolon associated with tuber formation. Question marks
indicate uncertain interactions/mobility or yet-to-be-identified interactors.
Note
: no differentiation has been
made between protein and gene in this figure.
same allele also phenocopied a number of other
characteristics of early potatoes such as drastic-
ally reduced life cycle and more bushy growth
habit (Kloosterman
et al.,
2013). The
StCDF1
gene was identified after fine mapping of the
earliness QTL on chromosome
5.
The wide variation
in plant earliness in modern potato cultivars
appears thus to be a result of allelic variation at a
single locus. This is likely to be due in part to the
selection for
StCDF1
genes that evade the circadian
rhythm control by StFKF1, as demonstrated in a
genome-wide association study in
83
diverse
tetraploid genotypes where a unique and very
high association was found between earliness
and the CDF1 locus on chromosome
5
(Klooster-
man
et al.,
2013).
The similarity between the flowering and
tuberization regulation pathways makes it
likely that signaling cascade between the clock
output proteins and FT in potato has been re- or