Biomedical Engineering Reference
In-Depth Information
A
A
T
T
C
A
T
G
A
C
T
G
A
A
T
T
C
A
T
G
T
A
G
T
A
C
A
C
T
G
T
G
A
C
T
T
A
A
G
T
A
C
T
G
A
C
T
T
A
A
Restriction enzyme
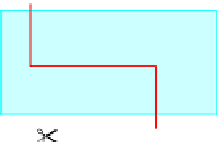
FIGURE 14.8
An illustration of the effect of restrictive enzyme.
foreign genes into the cells. To prepare donor DNA and vector for recombination, special
enzymes are required. A wide variety of restriction endonucleases and ligases are available
and are described below.
14.4.5.1. Restriction Enzymes
These are enzymes that cut DNA at specific sites. They are properly called restriction
endonucleases because they cut the bonds in the middle of the polynucleotide chain. For
example, EcoRI is a restriction endonuclease from
E. coli
. Some restriction enzymes cut
straight across both chains, forming blunt ends, but most enzymes make a staggered cut
in the two strands, forming sticky ends (
Fig. 14.8
).
The cut ends are “sticky” because they have short stretches of single-stranded DNAwith
complementary sequences. These sticky ends will stick (or anneal) to another piece of DNA
by complementary base pairing, but only if they have both been cut with the same restriction
enzyme. Restriction enzymes are highly specific and will only cut DNA at specific base
sequences, 4
e
8 base pairs long, called recognition sequences.
Restriction enzymes are produced naturally by bacteria as a defense against viruses (they
“restrict” viral growth), but they are enormously useful in genetic engineering for cutting
DNA at precise places (molecular scissors). Short lengths of DNA cut out by restriction
enzymes are called restriction fragments. There are thousands of different restriction
enzymes known, with over a hundred different recognition sequences. Restriction enzymes
are named after the bacteria species they came from, so EcoR1 is from
E. coli
strain R and Hin-
dIII is from
Haemophilus influenzae
. Some examples of restriction enzyme and their sources are
shown in
Table 14.3
.
14.4.5.2. DNA Ligase
This enzyme repairs broken DNA by joining two nucleotides in a DNA strand. It is
commonly used in genetic engineering to do the reverse of a restriction enzyme, i.e. to join
together complementary restriction fragments.
The sticky ends allow two complementary restriction fragments to anneal, but only by
weak hydrogen bonds, which can quite easily be broken, say by gentle heating. The backbone
is still incomplete.
DNA ligase completes the DNA backbone by forming covalent bonds. Restriction
enzymes and DNA ligase can therefore be used together to join lengths of DNA from
different sources (
Fig. 14.9
).
























































Search WWH ::

Custom Search