Biology Reference
In-Depth Information
7.4.2 CsgA Self-Assembly: Insights from
in vitro and in
silico
Research
Studies of the self-assembly of pure CsgA
proved that it forms
amyloids that share common features with “classic” amyloidogenic
disease-associated proteins. CsgA was shown to display nucleation-
dependent polymerization kinetics with lag, growth, and stationary
phases.
in vitro
32
As viewed by electron microscopy, �ibre formation occurs
already two hours after CsgA puri�ication. The �ibre formation
process was demonstrated to be accelerated by the presence of
preformed CsgA �ibres signi�icantly shortening the lag phase (process
known as “seeding”).
32
A
A
(a)
S
ELNIY
Q
Y
GGG
N
S
A
LAL
Q
TDARN
S
DLTIT
Q
H
GGG
N
G
A
DVG
Q
-GSDD
S
SIDLT
Q
R
G
F
G
N
S
A
TLD
Q
WNGKN
S
EMTVK
Q
F
GGG
N
G
A
AVD
Q
-TASN
S
SVNVT
Q
V
G
F
G
N
N
A
TAH
Q
Y
Hexarepeat
S
ELNIY
Q
Y
GGG
N
S
A
LAL
Q
TDARN
S
DLTIT
Q
H
GGG
N
G
A
DVG
Q
-GSDD
S
SIDLT
Q
R
G
F
G
N
S
A
TLD
Q
WNGKN
S
EMTVK
Q
F
GGG
N
G
A
AVD
Q
-TASN
S
SVNVT
Q
V
G
F
G
N
N
A
TAH
Q
Y
Hexarepeat
C5a
C5b
C5c
C5d
C5e
C5a
C5b
C5c
C5d
C5e
(b)
B
B
6WUDQG
ORRSVWUDQG
motif
6WUDQG
ORRSVWUDQG
motif
3DUDOOHO
KHOL[
model
3DUDOOHO
KHOL[
model
Figure 7.3
CsgA internal homology and structural model. (a) Internal
alignment of the CsgA C-terminal domain. Conserved residues
within repeating units (C5a-e) are coloured in red (polar),
orange (glycines), blue (aromatics), and green (alanines). (b)
Sequence corresponding representation of predicted stand-
loop-strand motif. The assumed folding of the monomer into
the parallel
-helix structure is illustrated. See also Colour
Insert.
Interestingly, the structure of CsgA monomers after secretion was
natively unfolded.
However, using A11 conformational antibodies
that speci�ically recognize the transient species of amyloids
4,32
33
(such
as A
and IAPP), transient intermediate-like structures could also
be detected.
32
These were already present in the very beginning of
























































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































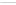
























































































































































































































































































































































































































































































































































































































































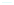













































































































Search WWH ::

Custom Search