Biology Reference
In-Depth Information
Appendix: Assays for
Marker-Assisted Selection
Corky Root
The assay was developed to distinguish alle-
les of the
SCO07
marker. This marker is
linked to the
cor
gene located at LG 3
(Moreno-Vazquez et al. 2003). The
SCO07
marker sequence from resistant cv. Green
Lake (AY207423) and susceptible cv. Diana
(AY207424) were downloaded from the NCBI
database
(http://www.ncbi.nlm.nih.gov).
The
480bp-long fragment from cv. Green Lake dif-
fers from the 481bp-long fragment from cv.
Three high-resolution DNA melting (HRM)
assays were developed at the USDA-ARS in Sali-
nas, California, for detecting alleles associated
with resistance to corky root, lettuce dieback,
and LMV (Figure 14.3). All assays were devel-
oped for a LightScanner system (96-well tray)
and analysis of melting profiles with LightScan-
ner software v. 2.0.0.1331 (both from Idaho
Technology, Salt Lake City, UT).
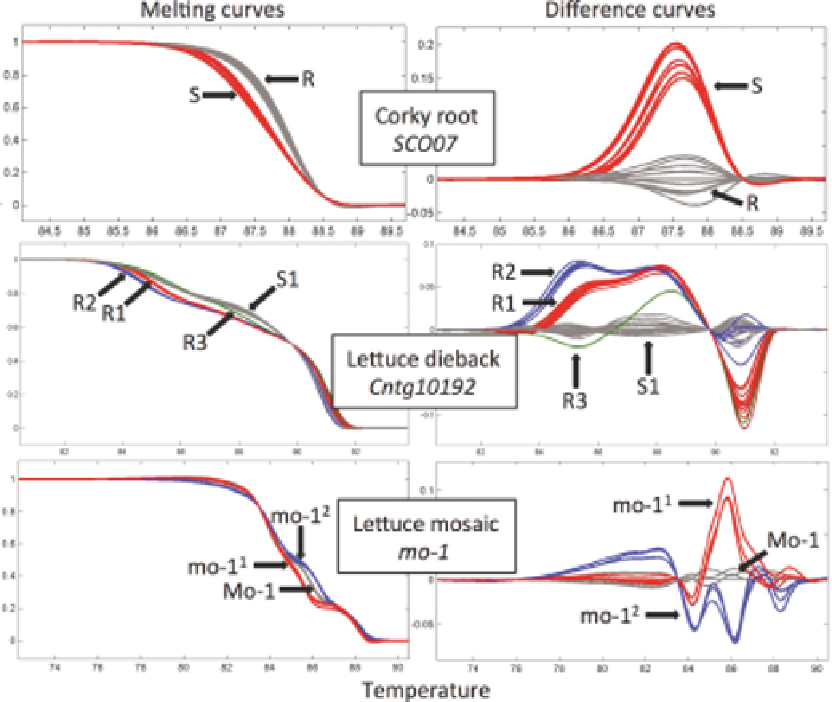
Fig. 14.3.
Examples of MAS with high-resolution DNA melting (HRM) assays. Top row - the
SCO07
marker linked to
the
cor
gene for resistance to corky root (R - allele linked to resistance; S - allele linked to susceptibility). Middle row - the
Cntg10192
marker linked to the
Tvr1
gene for resistance to lettuce dieback (R1 - allele linked to the 'Salinas' haplotype;
R2 - allele linked to the 'PI 491224' haplotype; R3 - allele linked to the 'UC96US23' haplotype; and S1 - allele linked to
the susceptible haplotype). Bottom row - alleles of the
mo-1
gene for resistance to lettuce mosaic (
Mo-1
is the susceptible
allele,
mo-1
1
and
mo-1
2
are resistant alleles). The
mo-1
assay is based on an unlabeled probe. In each assay only melting
curves of homozygous genotypes are shown, however heterozygous genotypes were also identified by HRM. For a color
version of this figure, please refer to the color plate.
Search WWH ::

Custom Search