Biology Reference
In-Depth Information
These results are entirely consistent with the earlier observations after TbrPDEB
knockdown with tetracycline-inducible RNAi (Oberholzer et al.
2007
).
These results provide pharmacological validation of the B class of kinetoplastid
phosphodiesterases as promising drug targets.
3.2 Crystal Structure of Leishmania major Phosphodiesterase
B1 and Implications for Selectivity of Parasite PDE Inhibitors
3.2.1 Structural Similarity and Variation Between
LmjPDEB1 and Human PDEs
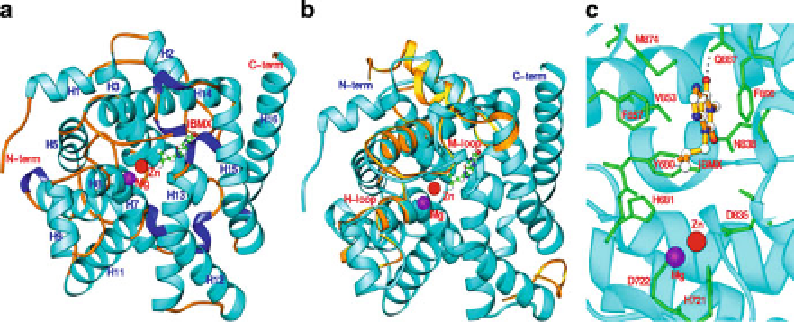
The structure of the catalytic domain of LmjPDEB1 (residues 582-940) in complex
with IBMX is the first and only structure of a parasite PDE determined to date
(Wang et al.
2007
). The catalytic domain contains 2 divalent metal ions and 16
a
-helices folded in an overall architecture similar to that of human PDEs (Wang
et al.
2007
; Ke and Wang
2007
). The superposition revealed four regions that exhi-
bit significant differences between the structures of LmjPDEB1 and human PDEs
(Fig.
6
). The most significant differences are associated with the H- and M-loops,
which have positional shifts of as much as 3
˚
for their C
a
atoms, about twice
the overall root-mean-squared deviation between the structures of LmjPDEB1 and
human PDEs. Since the H- and M-loops are directly involved in interaction with
inhibitors (Huai et al.
2004
; Wang et al.
2006
; Ke and Wang
2007
), their significant
positional shifts most likely have pharmacological implications, but further char-
acterization is needed to understand their exact roles.
Fig. 6 The structure of LmjPDEB1 (Wang et al.
2007
). (a)
Ribbon diagram
of the catalytic
domain of LmjPDEB1 in complex with the nonselective inhibitor, IBMX. (b) Comparison of
LmjPDEB1 (
cyan ribbons
) with PDE4D2. Only PDE4D loops (
gold ribbons
) with large positional
differences are shown. (c) IBMX binding at the active site of LmjPDEb1. The
dotted line
represents the hydrogen bond between IBMX and the side chain of glutamine