Biology Reference
In-Depth Information
Erk
PKA
Long
Catalytic
UCR1
UCR2
4D3/4/5/7/8/9; 4B1/3/4;
4A4/5/8/10/11; 4C1/2/3
Short
4D1; 4B2
Super-short
4D2/6; 4B5; 4A1
High Affinity Rolipram Binding Site
Catalytic Domain
PDE4A
PDE4B
PDE4C
PDE4D
Short
Super-short
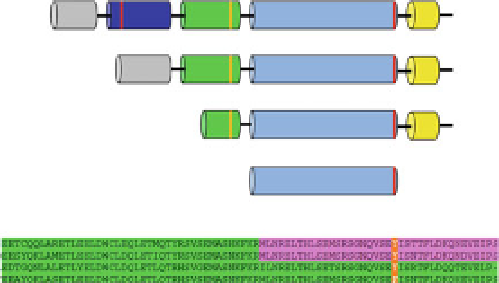
Fig. 1 Depiction of long, short and supershort splice isoforms of PDE4. Long isoforms of PDE4
contain UCR1(
blue
) and UCR2 (
green
) regulatory domains. The catalytic domain and C-terminal
sequence is identical across splice isoforms. Sites of protein kinase A (PKA) phosphorylation on
UCR1 (Ser54) and Erk phosphorylation on the catalytic domain (S579) are indicated. High-affinity
rolipram binding requires the C-terminal region of UCR2 (
pink
). UCR2 in long and short isoforms
of PDE4 contains amino acids 134-222. UCR2 is truncated to amino acids 167-222 in supershort
isoforms
Table 1 PDE4 UCR 2 gating elements are conserved across species
Human PDE4D
191 Asn-Gln-Val-Ser-Glu-Phe-Ile-Ser-
Asn-Thr-Phe-Leu-Asp 213
Human PDE4B
Asn-Gln-Val-Ser-Glu-Tyr-Ile-Ser-
Asn-Thr-Phe-Leu-Asp
Drosophila
(SwissProt Q9W4S9)
Asn-Gln-Ile-Ser-Glu-Tyr-Ile-Cys-
Ser-Thr-Phe-Leu-Asp
C.elegans
(SwissProt Q22000)
Thr-Gln-Val-Ser-Lys-Phe-Leu-Ile-
Thr-Thr-Tyr-Met-Asp
Key contact residues
----Gln----------Phe------------
-----Phe--------
The PDE4 UCR2
a
-helical gating sequence is shown for human PDE4D, Human PDE4B,
Drosophila melanogaster
(Chen et al.
1986
), and
Caenorhabditis elegans
(Charlie et al.
2006
).
Numbering of PDE4D UCR2 is based on the reference PDE4D3 isoform (GenBank AAA97892).
Gln192 is proposed to stabilize UCR2 in the closed conformation by forming a hydrogen bond
with Asn528 on the surface of the PDE4 catalytic domain. The Phe196/Tyr polymorphism affects
apparent
K
M
with respect to cAMP hydrolysis, perhaps through a hydrogen bond formed between
the Tyr on UCR2 and the 2
0
OH of AMP or cAMP. Phe201 fits into a hydrophobic pocket on the
surface of the PDE4 catalytic domain and has been shown by mutation analysis to be critical for
UCR2 capping (Burgin et al.
2010
)
a negative regulatory module, although structural information has been lacking
(Beard et al.
2000
; Houslay
2001
).
PDE4 is the major cAMP hydrolyzing enzyme in most cells, tissues, and
organisms. The complex pattern of splice isoforms allows PDE4 to be targeted to