Biomedical Engineering Reference
In-Depth Information
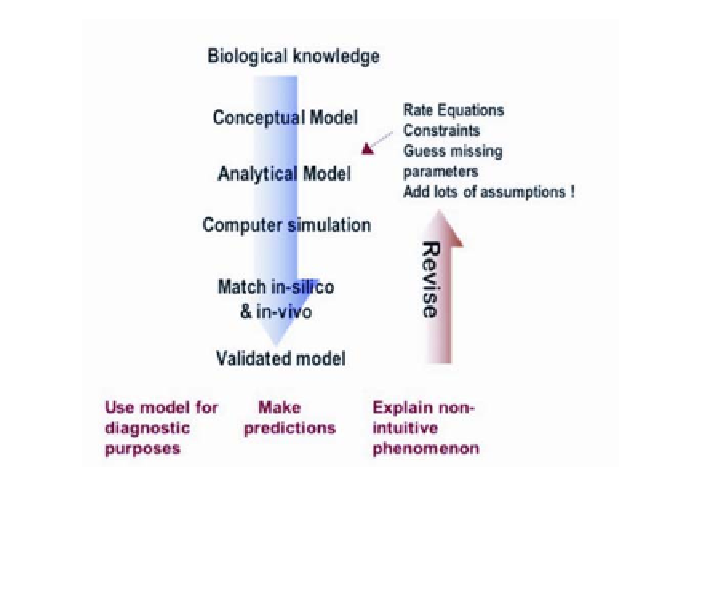
Figure 1
. Animating cellular processes
in silico
.
biology (1,2,22), but they realized the limitations of the classical systems ap-
proach. The underlying assumption was that physical laws were as applicable to
biological systems as to engineering systems. The dynamic and nonlinear nature
of living systems was not well understood, and researchers were comfortable in
viewing organisms as deterministic systems. Furthermore, the ease with which
measurements could be taken from physical systems was in stark contradiction
to biological systems, which posed major data-gathering challenges, leading to
problems in building precise analytical models. As a result, initial modeling ef-
forts were successful only to the extent of simulating cellular events, not in ex-
plaining fundamental principles. Following this, attempts were made to
construct toy models of biological systems under an assumption of steady-state
conditions. The most prominent of these were biochemical systems theory and
metabolic control theory. Another aspect that received more and more attention
was the appearance of patterns at different levels of biological complexity (24).
The last two decades have witnessed remarkable advancements in the field
of molecular and cellular biology. This has led to a better understanding of bio-
logical complexity, from molecules to organisms. The traditional and immensely
successful reductionist approach has resulted in the creation of huge databases.
However, to understand biology at its default (i.e., systems) level, data needs to
be woven into a system that not only portrays the known interactions but also