Biomedical Engineering Reference
In-Depth Information
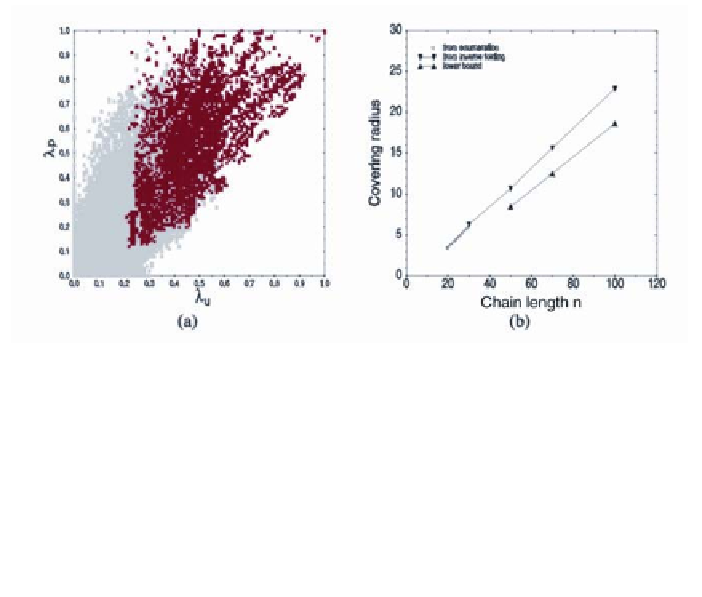
Figure 2
.
Neutral Networks and Shape Space Covering
. (
a
) Neutral networks in an exhaustive
survey of the
GC
sequence space with length
n
= 30 (43) are fragmented (light grey) if the
fractions M
u
and M
p
of neutral mutations in the unpaired and paired parts of the sequence are
below a threshold value. Above the threshold the neutral networks consist of one to four con-
nected components. The fragmentation of the single connected component into a small number
of (barely) separated subsets can be explained by the details of an energy-based folding model;
see (124). (
b
) The shape space covering radius
d
cov
scales linearly with the chain length
n
with a
slope K 1/4. Data are taken from (43).
thus exhibits a remarkable combination of robustness and fragility, typical for
many complex systems (see also Part II, chapter 5, by Krakauer, this volume).
Neutral networks and shape space covering are emergent properties in this set-
ting.
The set of nodes of the neutral network
f
-1
(
s
) is embedded in a compatible
set
C
(
s
) that includes all sequences that can form the structure
s
as suboptimal
or
minimum free energy conformation
f
-1
(
S
) I
C
(
s
). Sequences at the intersection
C
(
s
')
C
(
s
'') of the compatible sets of two neutral networks in the same se-
quence space are of actual interest because these sequences can simultaneously
carry properties of the different RNA folds. For example, they can exhibit cata-
lytic activities of two different ribozymes at the same time (120). The intersec-
tion theorem (108) states that for all pairs of structures
s
' and
s
'' the intersection
C
(
s
')
C
(
s
'') is always non-empty. In other words, for each arbitrarily chosen
pair of structures there will be at least one sequence that can form both. If
s
' and
s
'' are both common structures, bistable molecules that have equal preference for
both structures are easy to design (25,53). A particularly interesting experimen-
tal case is described in (120).
At least features (i), (ii), and (iv) of the neutral networks of RNA seem to
hold for the more complicated protein spaces as well (3,4) (see, e.g. (71) for
experimental data).