Image Processing Reference
In-Depth Information
a
b
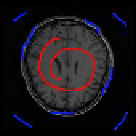
Fig. 9.8
Example of labels in axial plane. (a) Selected slice. (b) labels in image. Red pixels
indicate desired objects while blue pixels denote background.
issue is that the local errors can be re-segmented as new object. The additional seeds
throughout the computation do not increase the whole working time or only a little
time cost. In realistic systems, user can set data boundary to decrease re-computing
time largely. Here, eye tissues need to be erased (see Figure 9.10). The validation
also indicates that computing time has no explicit relations with slice and seed num-
bers when the seed number is less than 1/10 of total pixels. The segmented tissues
data can be visualized with Marching Cubes [20] [24] and Ray-casting [17] to ren-
der (see Figure 9.11).
It is a heavy burden for computing on the whole volumetric data set, so accelera-
tion is crucial for the applications based on CA.
9.3.3.2
Acceleration with Pyramid
In CA evolution, time cost is almost direct ratio to data magnitude, Gaussian and
Laplacian pyramid method [4] can be used to produce re-sampled layer data so as to
accelerate segmentation. The layer number is specified by user according to interac-
tion precision. In higher level layer, pre-segmentation is performed to get a coarse
a
b
c
d
e
f
g
h
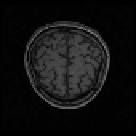
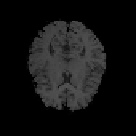
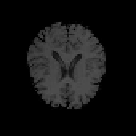
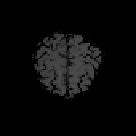
Fig. 9.9
Results of 3D CA segmentation. (a) - (d) Samples of original series. (e) - (h) Seg-
mented tissues of brain (location = 20, 25, 28, 40).





Search WWH ::

Custom Search