Chemistry Reference
In-Depth Information
another transcript. In any case, the approach was not appropriate for studying
the dynamics of a single RNA molecule. The key for this advance came from the
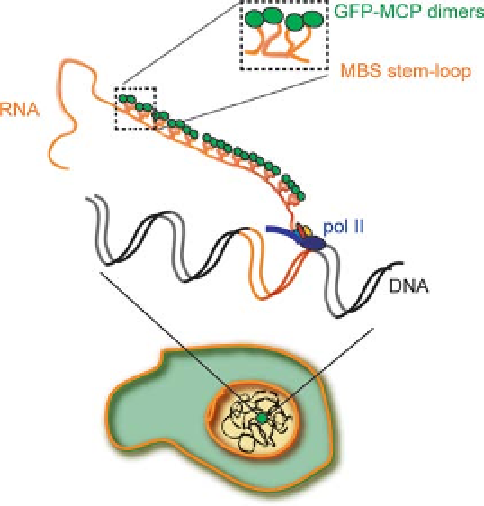
bacteriophage MS2 coat protein (MCP) coupled with FP tagging [21]. This phage
protein is an RNA-binding protein recognizing speci
cally a distinctive binding
site on a stem
-
loop folded RNA (Figure 8.1). The high af
nity interaction (
1 nM)
between the stem
-
loop and the phage protein make this method highly speci
c.
Since twoMCPs bind each stem
-
loop as dimers, the insertion of several MS2 binding
sites (MBS) into the target gene will recruit multiple
<
orescent tagged MCPs on a
single molecule (Figure 8.1), providing a powerful system for the detection of single
mRNPs distinguishable from the GFP-MCP background.
Developing this system in yeast [21] and in human cells [22, 23] made it possible to
probe RNAexpression of a speci
c sequence. A recent study focused on the synthesis
of a speci
c gene array of MBS containing-transcripts by pol II and demonstrated the
advantages of the MS2 system in mammalian cells [24]. In this work, integration of
results obtained by FRAP, photoactivation, mathematical modeling and computation
analysis allowed the quanti
cation of the in vivo dynamics and kinetics of pol II
transcription.
Coupling this method with other emerging tools like Fluorescence Correlation
Spectroscopy (FCS), ameans of resolvingmolecular events within rapid time frames,
for example the dynamics of the speci
c steps of transcription or for splicing, will be
likely to yield valuable kinetic data.
Figure 8.1 Schematic representation of a
specific RNA labeled with theMS2 system. Upon
stable insertion of the modified gene in the cell,
its active transcription site will be visualized by
fluorescence due to the binding of the GFP-MCP
dimers at the MCB stem
simplicity only a single transcribing polymerase
is depicted. Multiple active polymerases at the
site will increase the RNA production therefore
the intensity, allowing the site to be detected over
the signal background.
loop regions. For
-