Biology Reference
In-Depth Information
C. Inverse PCR
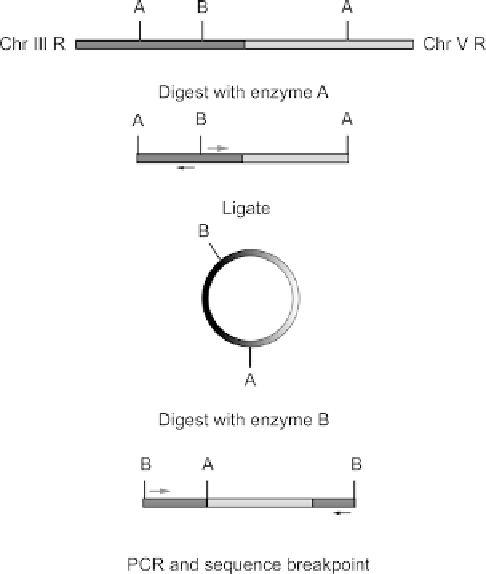
In the case of a balancer in which the location of one breakpoint has been mapped
to a small region of the genome that information can be used to perform inverse PCR
of the rearranged chromosomes. This approach will identify the remaining break-
point position. In the case for eT1, which breaks in unc-36 (III), PCR across the 7 kb
long gene was used to locate the chromosome III breakpoint to a 450 bp interval.
Inverse PCR was subsequently used to identify the sequence of the fusion between
chromosome III and chromosome V (
Zhao et al., 2006
)(
Fig. 9
). This information
was useful for developing a PCR assay to rapidly identify the eT1 chromosomes in
genetic crosses (
Zhao et al., 2006
).
D. Whole Genome Sequencing
Advances in sequencing technology have made whole genome sequencing experi-
ments feasible. There are a growing number of C. elegans whole genome sequences
available (
Hillier et al., 2007, 2008; Rose et al., 2010
), in addition to the canonical
assembled and aligned sequence in WormBase (
Harris
et al., 2010
)(
www.
Fig. 9
iPCR scheme is used to isolate eT1 breakpoint. See text for details.
Search WWH ::

Custom Search