Biology Reference
In-Depth Information
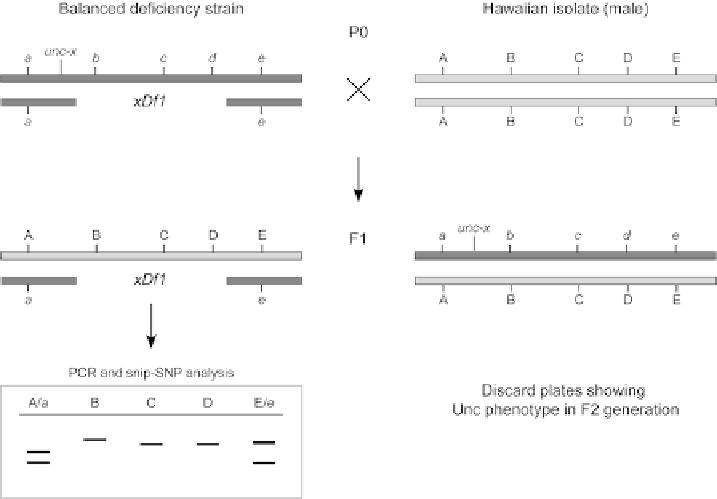
Fig. 7
SNP mapping strategy for deficiency mapping. A strain heterozygous for the deficiency (in this
hypothetical example xDf1) to be characterized and a balanced visible phenotypic marker is mated to the
Hawaiian isolate strain. F1 progeny arising from this cross are individually picked and the resultant F2
animals scored for the presence of the visible phenotype (mut-x). F2 populations not displaying the mut-x
phenotype (and therefore maintaining xDf1) are cultured, genomic DNA is extracted and analyzed using
appropriate snip-SNPs.
A crossing scheme for SNP mapping a deficiency is shown in
Fig. 7
. The balanced
Df strain is crossed to Hawaiian males. The F1 progeny will be heterozygous for
Bristol and Hawaiian chromosomes and receive either the Df or the balancer chro-
mosome. If the balancer is marked, it is possible to enrich for the heterozygotes.
DNA from the F2 progeny is extracted and used as template in PCR amplification. It
is important to prepare the DNA from the F2 generation to ensure that the viable
Hawaiian chromosome does not outcompete the deficiency chromosome
(
Kadandale et al., 2005
). Since the DNA template can be used for multiple PCR
reactions a strategy can be used to first roughly position the deficiency with broad
markers and then use subsequent fine mapping in increments until SNPs are
exhausted or breakpoints located to a sufficient resolution.
1. Advantages and Disadvantages
SNP analysis has several advantages over more traditional mapping techniques.
The approach is rapid as a single genetic cross and DNA extraction followed by PCR
restriction analysis can be done in a less than two weeks. No specific equipment or
Search WWH ::

Custom Search