Biology Reference
In-Depth Information
of novel small RNAs, as it is not limited by a priori sequence information. For
example,
Ruby et al. (2006)
applied 454 pyrosequencing to small RNAs expressed in
mixed-staged C. elegans and identified the class of 21U-RNAs and additional
previously unknown miRNAs (
Ruby et al., 2006
). Our group also has used
Illumina Genome Analyzer for profiling dynamic miRNA expression in C. elegans
development and identifying novel miRNAs and 21U-RNAs (
Kato et al., 2009
).
Protocols for creating cDNA libraries of small RNAs commonly include size
fractionation of RNAs, 5
0
- and 3
0
-adaptor ligation, reverse transcription, and PCR
amplification (
Lagos-Quintana et al., 2001; Lau et al., 2001; Lee and Ambros,
2001
). These cDNA libraries are made amenable for deep sequencing by the inclu-
sion of appropriate primer sequences. Commercially available kit systems for pre-
paring cDNA libraries for deep sequencing also have been released. In the
approaches described below, we follow the manufacturer
'
s instruction in the
Illumina
'
s Small RNA Sample Prep Kit (catalog # FC-102-1009) for preparing
cDNA libraries and discuss the principles of deep sequencing of C. elegans small
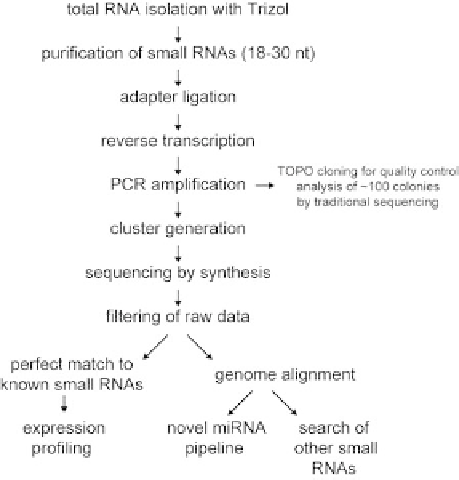
RNA populations based on our approach using Illumina Genome Analyzer, sum-
marized graphically in
Fig. 1
. These general principles are broadly applicable to
other platforms.
Fig. 1
An outline of deep sequencing of small RNA populations. See text for details.
Search WWH ::

Custom Search