Biology Reference
In-Depth Information
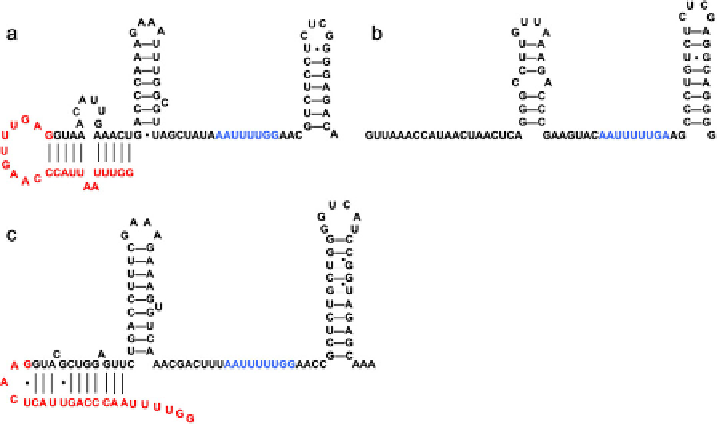
Fig. 3
(a) SL1 RNA predicted secondary structure. (b) SmY-1 RNA predicted secondary structure.
Other SmY snRNAs vary in sequence but retain this secondary structure. (c) SL2 RNA predicted
secondary structure. All the SL2 RNA variants also retain this secondary structure. In the SL RNAs,
the spliced leader sequence is shown in red. In all structures, the Sm-binding site is shown in blue. (See
color plate.)
thoroughly explored. Only deletions or substitutions in the loop immediately
upstream of the splice site resulted in constructs capable of rescuing rrs-1 lethality
(
Ferguson and Rothman, 1999
).
The Sm-binding domain is flanked by the second and third stem-loops. In vitro
studies have identified a region of the stem on the second stem-loop which, when
mutated, results in an SL RNA incapable of trans-splicing (
Denker et al., 1996;
Hannon et al., 1992
). These studies also demonstrated that the Sm-binding site is a
necessary component of a functional SL snRNP. Finally, the aforementioned in vivo
study showed that embryonic rescue is abolished when the transgenic SL1 RNA does
not contain this Sm-binding domain (
Ferguson et al., 1996
).
3. Catalysis of Trans-Splicing
Trans-splicing of a pre-mRNA depends on the presence of a 3
0
splice site upstream
from the coding sequence, without an accompanying 5
0
splice site upstream (
Conrad
et al., 1991, 1993a
). In addition to studies in C. elegans, much of the trans-splicing
mechanism has been elucidated from studies done in Ascaris extract. The trans-
splice site on the pre-mRNA is presumably bound by the U2AF and SF1/BBP
proteins and the U2 snRNP, as would occur during cis-splicing, since the consensus
sequences for cis- and trans-3
0
splice sites are the same (
Hollins
et al., 2005
).
Search WWH ::

Custom Search