Biology Reference
In-Depth Information
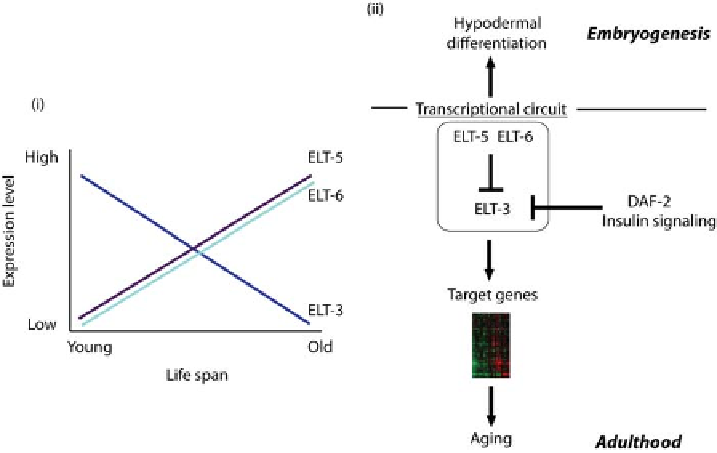
Fig. 5
Expression changes of elt-3/elt-5/elt-6 during normal aging and the model for this transcrip-
tional circuit. (i) During the normal aging process, the expression of the GATA transcription factors elt-5
and elt-6 increases. This represses the expression of the GATA transcription factor elt-3. (ii) The
transcriptional circuit consisting of elt-3, elt-5, and elt-6 regulates hypodermal differentiation during
embryogenesis and aging during adulthood. elt-5 and elt-6 promote normal aging by negatively regulating
elt-3 expression. ELT-3 regulates a group of age-dependent genes identified from DNA microarray
analysis. Consistent with a functional role in aging, loss of function of elt-3 suppresses the long-lifespan
phenotype of daf-2 mutants. The expression of elt-3 is repressed by the DAF-2 mediated insulin signaling,
indicating that this transcriptional circuit may also modulate the effects of the insulin signaling. (For color
version of this figure, the reader is referred to the web version of this topic.)
yeast system. If the two proteins physically interact, the two domains are brought into
close proximity, which triggers transcription of a reporter gene indicating that an
interaction has taken place. As an effort toward understanding protein-protein
interactions, a large-scale protein interaction (interactome) mapping project based
on Y2H screens is in progress (
http://interactome.dfci.harvard.edu/C_elegans/
)
(
Simonis et al., 2009
). This interactome map (Worm Interactome version 8) cur-
rently contains 3864 known binary protein-protein interactions. Through interac-
tome mapping, many interactive connections involving novel proteins have been
found between disparate biological processes in C. elegans (
Boulton et al., 2002;
Li et al., 2004; Reboul et al., 2003
). For example, using the Y2H screening strategy,
Tewari et al. (2004)
uncovered eight daf-7/TGF-
b
pathway modifiers that were not
identified by conventional means. They first employed six known components of the
daf-7/TGF-
b
pathway as bait for the first round Y2H screen using a C. elegans
cDNA library. In order to identify more novel interaction links, they conducted
the second round of Y2H screening using genes identified in the initial screen
Search WWH ::

Custom Search