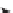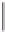Agriculture Reference
In-Depth Information
(A) Methane Consumption
Conventional
Early Successional
Mid-Successional
Mown Grassland
Deciduous Forest
15
10
5
0
(B) Stability of Methane Consumption
4
3
2
1
0
0
2 4 6 8 10
Methanotroph Species Richness (tRFLP OTUs)
Figure 6.4
. The number of bacterial species in soil that oxidize methane (methanotrophs) is
directly related to both the rate (A) and stability of methane consumption (B) at KBS LTER.
Methanotroph species richness is based on the number of Operational Taxonomic Units
(OTUs) defined by tRFLP analysis of methane (CH
4
) monooxygenase, the first enzyme in the
CH
4
oxidation pathway. Methane fluxes were estimated
in situ
using chambers placed over
the soil. The temporal stability of CH
4
consumption rates is expressed as the coefficient of
variation (CV) over the annual period of measurements. Redrawn from Levine et al. (2011).
Nitrate
Nitrite
Nitric oxide
Nitrous oxide
Dinitrogen
NO
3
-
NO
2
-
NO
N
2
O
N
2
Gene
:
Enzyme:
nor
nitric oxide
reductase
nos
nitrous oxide
reductase
nar
nitrate
reductase
nir
nitrite
reductase
Figure 6.5
. The bacterial denitrification pathway, including metabolic intermediates, essen-
tial genes and enzymes. A key enzyme in denitrification, producing the first gaseous metabo-
lite, is nitrite reductase (
nir
) encoded by the
nirK
or
nirS
genes. Both genes have been used
to survey the diversity of denitrifying bacteria.