Agriculture Reference
In-Depth Information
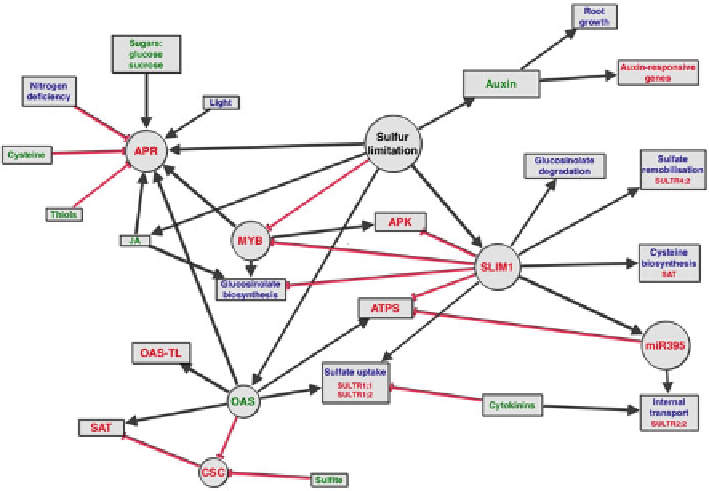
Fig. 3.5 Current understanding of the regulatory network of sulfate metabolism in
Arabidopsis
thaliana. Black arrows
indicate positive regulation (induction) and
red arrows
indicate negative
regulation (repression). The
grey circles
correspond to the key regulatory factors of the pathway,
grey boxes
correspond to all the other components involved in regulation.
Blue
colour indicates
processes,
red
- genes and proteins,
green
- metabolites. Abbreviations:
APR
APS reductase,
MYB
MYB factors,
APK
APS kinase,
SLIM1
Sulfur Limiting factor 1,
SULTRs
sulfate transporters,
SAT
serine acetyltransferase,
ATPS
ATP sulfurylase,
OAS-TL
OAS(thiol)lyase,
OAS O
-acetylserine,
CSC
cysteine synthase complex
Regulation on the Level of Sulfate Uptake and Transport
The efficient acquisition of sulfate from the soil and its distribution in the plant is of
great importance especially under sulfur limiting conditions. In a number of studies
it was shown that the rate of sulfate transport during low sulfate supply is driven
mainly by the regulation of the two high-affinity sulfate transporter genes,
SULTR1;1
and
SULTR1;2
(Shibagaki et al.
2002
; Takahashi et al.
2000
; Vidmar
et al.
2000
; Yoshimoto et al.
2002
). The study of promoter-reporter constructs
indicated that both are regulated in response to sulfate nutrition (Maruyama-
Nakashita et al.
2004a
). Further studies led to the identification of a transcription
factor, SLIM1 (sulfur limitation 1), which is responsible for the regulation of sulfate
uptake and metabolism during insufficient sulfur supply (Maruyama-Nakashita
et al.
2006
) (Fig.
3.5
). S
lim1
mutants showed about 30 % reduction in root length
and a 60 % decrease of sulfate uptake rates in sulfur-limiting conditions. The
SLIM1 transcription factor belongs to the family of ethylene insensitive-like
(EIL) transcription factors, from which EIL3 has a specific function in the