Agriculture Reference
In-Depth Information
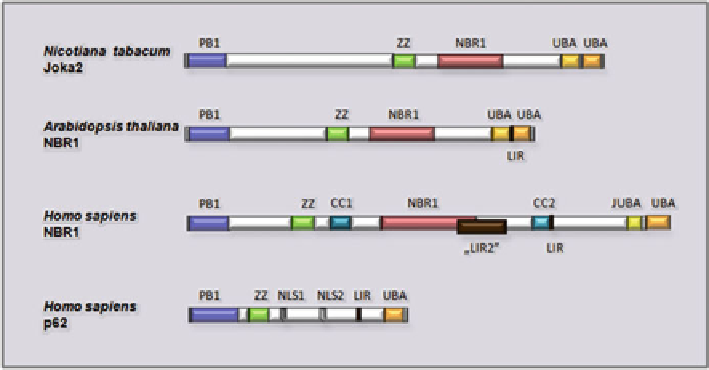
Fig. 7.6 The proteins are drawn to scale. The PB1, ZZ, NBR1, UBA1 and UBA2 domains are
marked. The experimentally verified nuclear localization signals (
NLS
) and mapped LIR motifs
are shown
PB1 Domain
The PB1 domain is a well-known interaction module, highly conserved among
animals, plants, fungi and amoebae (Sumimoto et al.
2007
) that can interact with
various proteins by creating salt bridges between basic and acidic residues located
on the PB1 domain to modify their functions. The PB1 domains are present in
nearly 200 proteins participating in diverse biological processes in all eukaryotes
(Letunic et al.
2002
). The PB1 domains could be classified into three groups based
on structure. Type-A is represented by PB1 domains possessing acidic OPCA
motif. Type-B includes PB1 domains with lysine residue/s on the first beta strand
carrying basic charge. The third group contains PB1 domains with both acidic and
basic clusters (Type-A+B).
The PB1 domain of human NBR1 has been classified as Type-A (Muller
et al.
2006
). No homodimers can be formed by this domain, while the PB1 domain
of human p62 is classified as Type-A+B, having both acidic and basic clusters, and
is involved in the formation of p62-p62 homodimers and in p62-NBR1
heterodimers. Similarly to p62, AtNBR1 (NBR1 from
Arabidopsis
) and Joka2
(a tobacco homologue) have been shown to form homodimers through the PB1
domain (Svenning et al.
2011
; Zientara-Rytter et al.
2011
).
Oligomerisation of the selective autophagy cargo receptors is crucial for degra-
dation process since mutants of p62 and AtNBR1, with substituted amino acids that
are particularly important for polymerisation, lost the ability to aggregate and are
not degraded by autophagy. Consequently, the human NBR1, possessing only an
OPCA motif is not able to self-interact through PB1 and an additional CC (coiled-
coil) region is required for self-oligomerisation.