Information Technology Reference
In-Depth Information
A
B
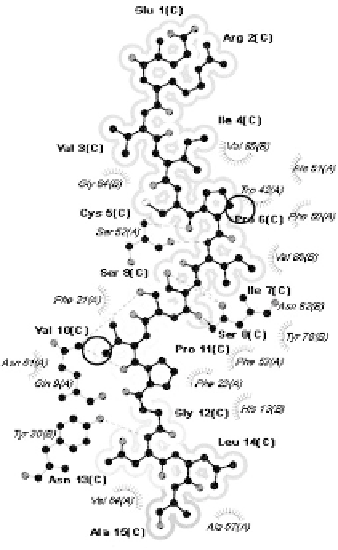
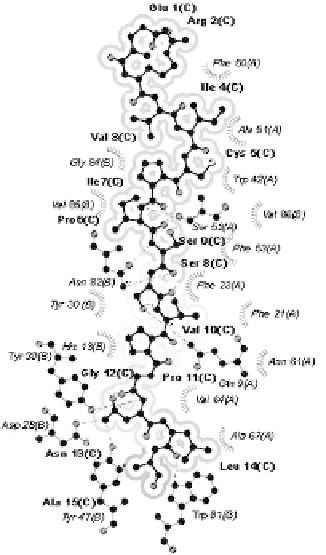
Fig. 3.
Comparison of Dsg3 963-977 (ERVICPISSVPGNLA) peptide within the
binding grooves of associated and non-associated alleles at 4.00Å. (A) Strongly
associated allele *0402. No atomic clash is detected in the modeled peptide-MHC
complex. (B) Non-associated allele *0406. Buried peptide residues are shaded grey
(in increasing density) and regions of atomic clash occurring between peptide and
pocket residues are circled in black.
3.6 Available Resources
A comprehensive dataset to facilitate the sequence-structure-function mapping in
peptide binding by MHC receptors is essential for structural analysis and
development of predictive algorithms in computational immunology. Listed in Table 1
below are some pMHC structural databases that are freely available for use or
download.