Information Technology Reference
In-Depth Information
root-mean-square deviation RMSD (Tong, Tan, and Ranganathan 2004) from the
experimental crystal structure. This chapter introduces the use of three-dimensional
experimental and modeled structures for peptide-MHC (pMHC) prediction. First, we
provide a brief description of the structural characteristics of pMHC complex
followed by an introduction on the use of structural descriptors to characterize
pMHC interfaces. Following this, we introduce the use of protein structure prediction
techniques to generate models of pMHC complexes. Finally, we cover some
available pMHC structural resources from the Internet.
3.2 Structural Features of MHC Peptides
Two main classes of MHC molecules, class I and class II, are identified that are
responsible for presenting epitopes to cytotoxic T cells (T
c
) and helper T cells (T
h
),
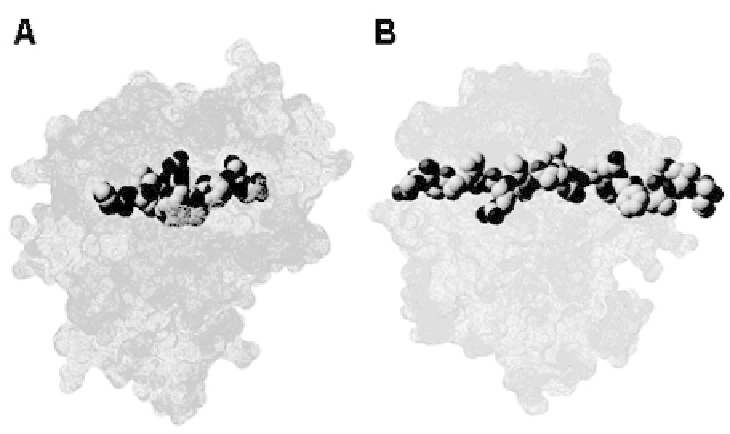
respectively. Each class has different binding characteristics: (i) class I ligands with a
typical length of between 8 and 12 amino acids are enclosed within the MHC
binding groove with both termini tethered toward the base of the binding groove and
the center loop bulges out to interact with T
c
(Fig. 1A); (ii) class ΙΙ ligands (Fig. 1B)
are more variable with both termini extending out of the groove thus permitting T
h
to
bind to ligands of a longer length (between 9 and 25 amino acids).
Fig. 1.
(A) Class I MHC-peptide complex. (B) Class II MHC-peptide complex.