Information Technology Reference
In-Depth Information
identified mutations, the process was very labor intensive and not cost effective. Hence,
we started to investigate other high-throughput mutation discovery systems.
A new mutation discovery system, Temperature Gradient Capillary Electrophore-
sis (TGCE), became commercially available in 2002. The TGCE system detects the
difference of a heteroduplex DNA fragment from the homoduplex counterpart during
the capillary electrophoresis (Gao and Yeung 2000; Murphy, Hafez, Philips, Yarnell,
Gutshall, and Berg 2003). The formation of heteroduplex fragments by a point muta-
tion is schematically shown in Fig. 8.
By using the TGCE system as the primary screening method for identifying mu-
tations in the target sequences of the G1 genomic DNA archive, the ENU-based
gene-driven mutagenesis has become practicable. At present, the mutation discovery
rate is about 100 per year per TGCE system (Sakuraba
et al. 2005). Each mutation
corresponds to one knockout mouse line. Since it takes more than a year to character-
ize the phenotypes, the mutation discovery rate vastly exceeds the capacity of the
biological and functional analyses of the mutant mouse lines. The size of the RIKEN
frozen sperm archive is about 10,000 as shown above (Sakuraba
et al. 2005). In this
archive, 3 × 10
7
independent point mutations are preserved for the biological analy-
ses (see Eq. (1) in Section 10.3.3.1). This is a tremendous genetic resource for the
biological annotation of the genome function.
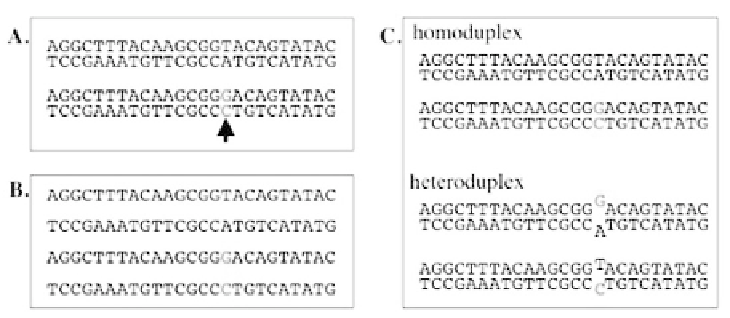
Fig. 8.
PCR fragments amplified from G1 mutant carrying an ENU-induced mutation and the
heteroduplex formation. (A) Design of PCR primers that amplify a target fragment of the G1
genome. When the target region of the G1 genome carries an ENU-induced mutation (arrow),
it appears only in the paternally inherited genome as heterozygote. (B) Heat denaturation of
PCR products makes all the fragments single stranded. (C) Renaturation of the heat-denatured
PCR products reconstructs the double helix structure and heteroduplex fragments that have a
mismatch-pairing at the site of the mutation. Theoretically the molar ratio is equal between
homoduplex and heteroduplex fragments.