Information Technology Reference
In-Depth Information
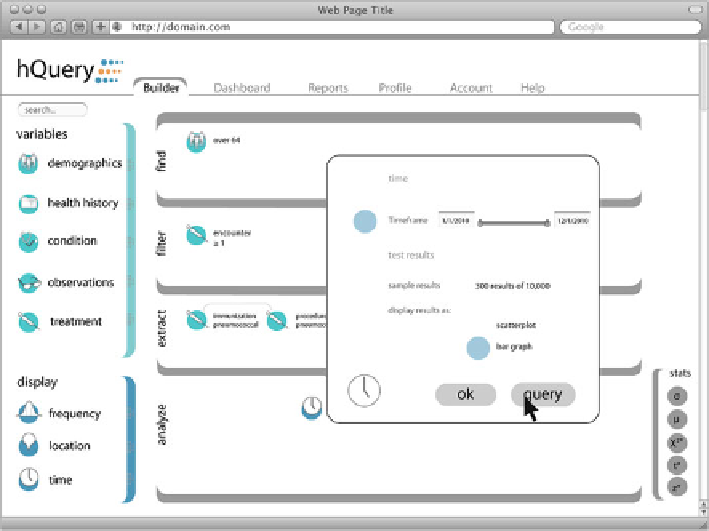
Fig. 7.1
hQuery Provides a Simple Web-based User Interface to Specify the Specific Questions
for which Answers are Sought
Informatics for Integrating Biology and the Bedside (i2b2)
is an NIH-funded
collaborative research project involving many of the major health and technology
institutions in Boston. Its core mission includes bringing together routine patient
data obtained in clinical care with the genome-wide measurements made of the cor-
responding patients. This is often expressed as combining genomic data with the
phenotypic data to gain a deeper understanding of the impact of genomic alleles
(variations).
The i2b2 analog to PopMedNet's Portal is called the i2b2 “Hive”. The Hive con-
sists of substitutable and interoperable software components called “Cells” and of a
persistent data storage called the Clinical Research Chart. Many of the cells have a
user level interface and are grouped in the “Workbench” where users can search the
i2b2 terminology, build and store queries with concepts from this terminology, and
visualize the resulting data. It has been used with reasonable success at the University
of Utah to test the applicability of an asthma exacerbation predication tool devel-
oped in Boston on the patient data resident in their institution. [
7
]
hQuery
is an open-source implementation of a framework for distributed query of
health data funded by ONC. For ease of implementation and use it emphasizes a
simple approach based on current web standards, as shown in Fig.
7.1
. Like the
other technologies, hQuery provides distributed query capabilities that maintain