Biology Reference
In-Depth Information
such maps based on information extracted from the scientific literature and/or
experimental data [72]. In this work, we adopted the framework implemented
by PathwayAssist [73, 75] for the construction of protein interaction maps. Path-
wayAssist effectively mines over 100,000 events of regulation, interaction and
modification between protein, cell processes and small molecules, thus allowing
the construction of complex interaction networks. It must be emphasized that al-
though the results might be sensitive to the specific software used, the essence of
the network properties is not. In order to computationally construct the interac-
tion network, the genes comprising the 9 most populated motifs (per the selection
procedure earlier outlined) when analyzed using Ariadne Genomics PathwayAs-
sist. The nodes that were selected were such that only direct connections between
them could be drawn, therefore establishing either direct links between nodes, or
indirect links using only nodes from the original set of informative genes.
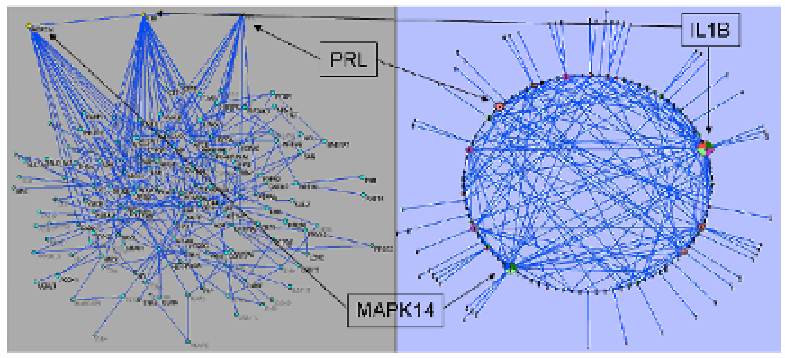
Despite the apparent complexity of the resulting network, Fig. 3.9, it is in-
teresting to realize the validation of the emergence of non-random connections as
previously speculated for protein interaction networks. The scale-free nature of
the resulting protein interaction network emanating from the analysis of informa-
tive genes identified by SLINGSHOTS a gene selection algorithm developed by
our group is further established by observing the distribution of arcs per node of
the network depicted in Fig. 3.10. Clearly, only a small fraction of nodes has a
characteristically large number of direct neighbors, which is an indication of the
emergence of a small number of highly connected nodes.
Fig. 3.9. Alternative representations of the general network topology of the protein interaction net-
work indicating the existence of highly connected hubs.
The scale-free character of the protein interaction networks is precisely what








Search WWH ::

Custom Search