Biology Reference
In-Depth Information
popular pages, then the overall structure of the network is scale free [62, 63]. The
presence of scale-free subnets suggests that the creation of individual pathways
is therefore an iterative process by which pre-existing pathways are augmented
and modified to provide an evolutionarily beneficial response. It would suggest
that biological mechanisms are not “irreducibly” complex and that the network
structure is consistent with an iterative evolutionary process.
We hypothesize that transcription factor binding sites which represent points
of intervention have the following qualities:
(1) They are localized to a high degree to a specific cluster or process
(2) They are prevalent to a high degree in spite of their selectivity
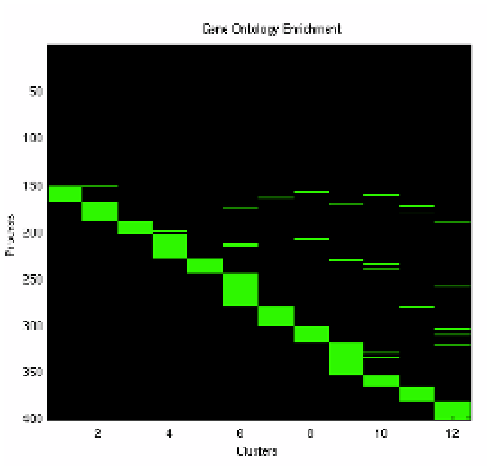
In Fig. 3.8, we show the relative localizations of the biological process ontolo-
gies into each individual clusters. What is evident is that given our clustering and
selection, we have isolated a set of genes which show a co-functional relationship
along with their co-expression. What this signifies is that each set of co-expressed
genes has a consistent scale-free network, and each of these scale free networks
appear to be regulating a specific but loosely independent set of biological pro-
cesses. The term loosely independent set is used to denote the fact that while
not all biological processes are localized to only one cluster, there is a predilec-
tion of ontologies to be located in a single cluster as evidenced by the diagonally
dominant plot shown in Fig. 3.8.
Fig. 3.8.
Biological process enrichment of the extracted genes. The processes have been thresholded
(
p<.
05).








Search WWH ::

Custom Search